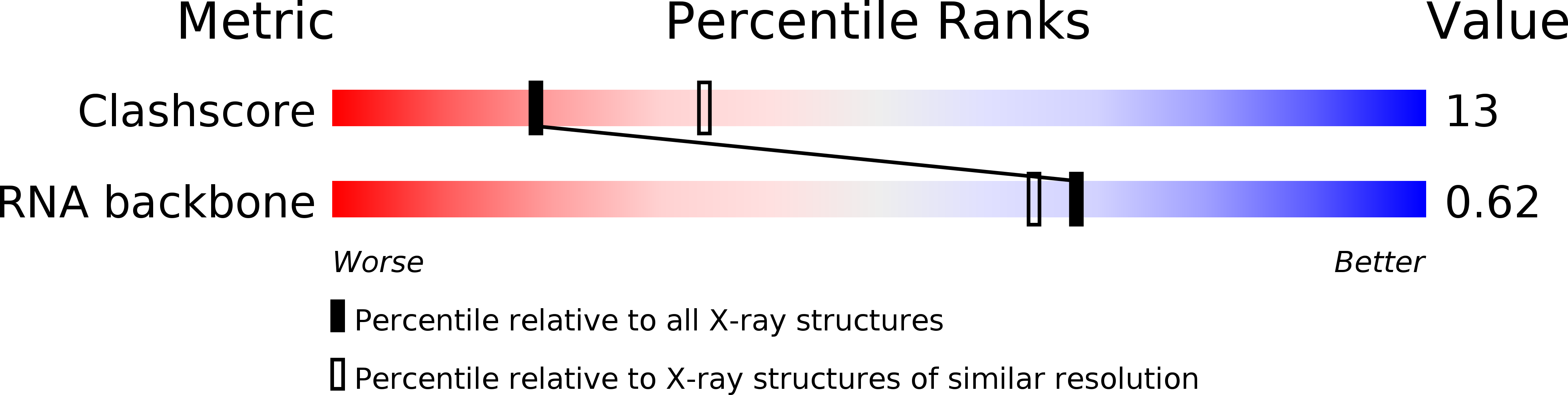Deposition Date
2002-08-20
Release Date
2002-09-06
Last Version Date
2024-02-14
Entry Detail
PDB ID:
1MHK
Keywords:
Title:
Crystal Structure Analysis of a 26mer RNA molecule, representing a new RNA motif, the hook-turn
Method Details:
Experimental Method:
Resolution:
2.50 Å
R-Value Free:
0.27
R-Value Work:
0.23
R-Value Observed:
0.24
Space Group:
C 1 2 1