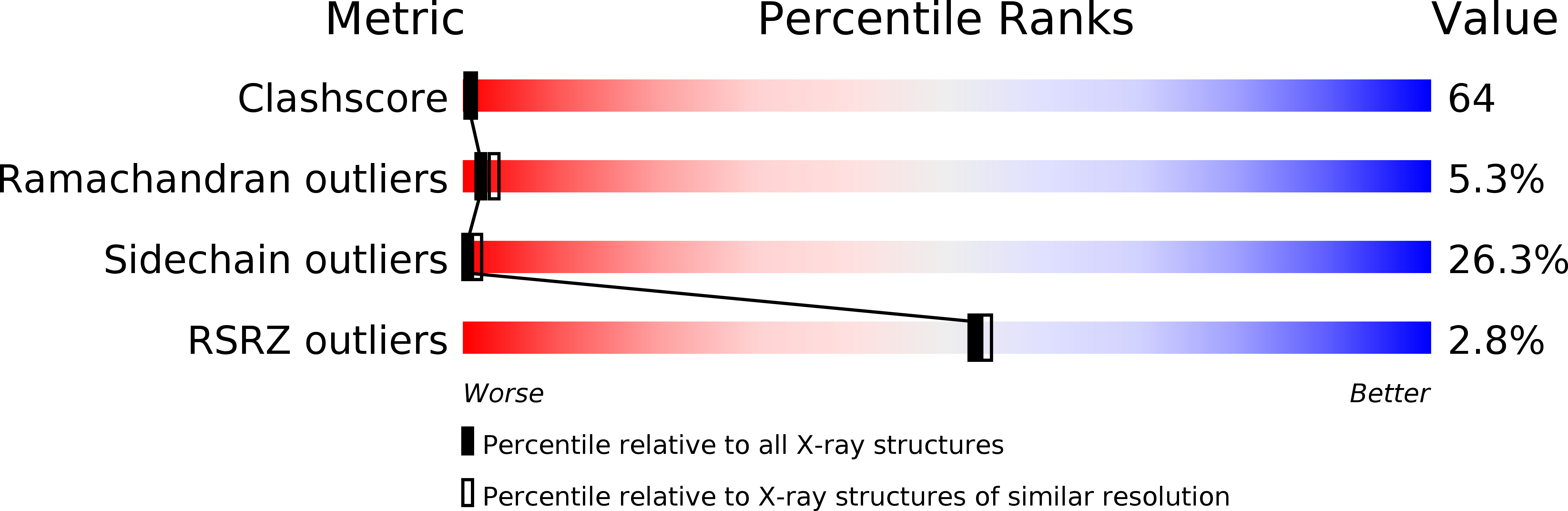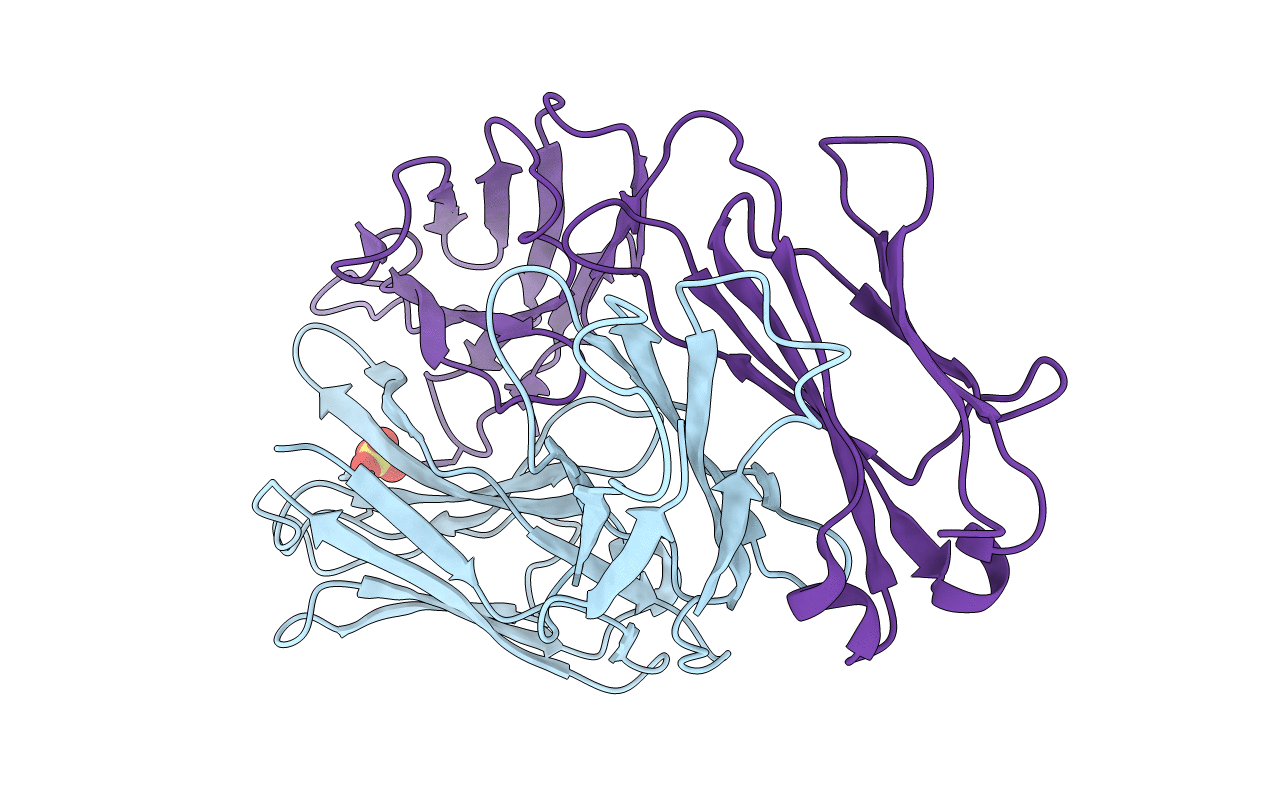
Deposition Date
1984-07-09
Release Date
1985-01-02
Last Version Date
2024-11-13
Entry Detail
PDB ID:
1MCP
Keywords:
Title:
PHOSPHOCHOLINE BINDING IMMUNOGLOBULIN FAB MC/PC603. AN X-RAY DIFFRACTION STUDY AT 2.7 ANGSTROMS
Biological Source:
Source Organism(s):
Mus musculus (Taxon ID: 10090)
Method Details: