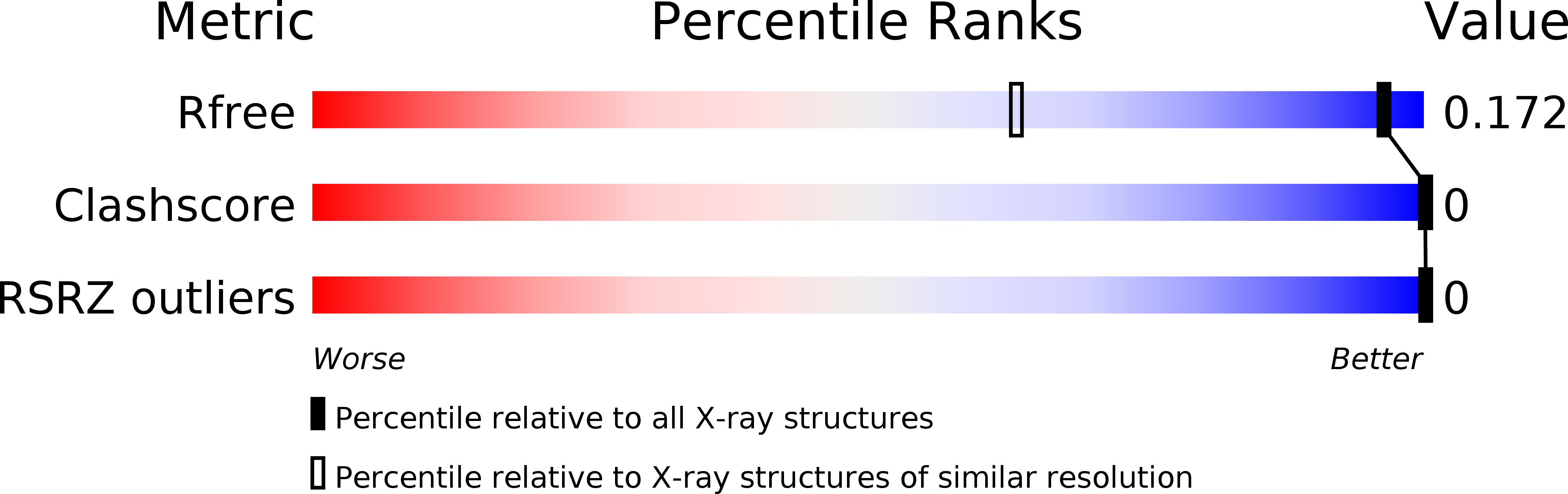Deposition Date
2002-07-18
Release Date
2003-01-07
Last Version Date
2024-04-03
Entry Detail
PDB ID:
1M77
Keywords:
Title:
Near Atomic Resolution Crystal Structure of an A-DNA Decamer d(CCCGATCGGG): Cobalt Hexammine Interactions with A-DNA
Method Details:
Experimental Method:
Resolution:
1.25 Å
R-Value Free:
0.18
R-Value Work:
0.16
Space Group:
P 43 21 2