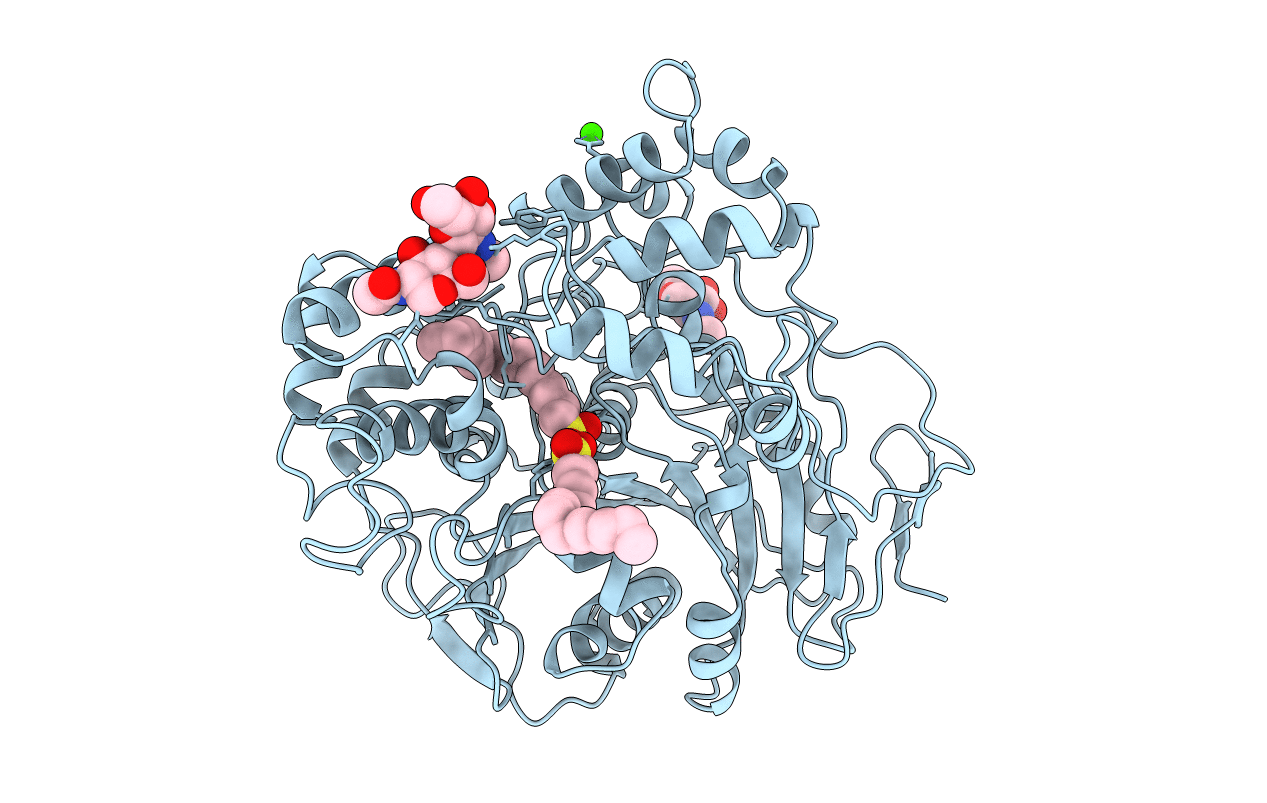
Deposition Date
1995-01-17
Release Date
1995-04-20
Last Version Date
2024-10-23
Entry Detail
PDB ID:
1LPP
Keywords:
Title:
ANALOGS OF REACTION INTERMEDIATES IDENTIFY A UNIQUE SUBSTRATE BINDING SITE IN CANDIDA RUGOSA LIPASE
Biological Source:
Source Organism(s):
Candida rugosa (Taxon ID: 5481)
Method Details:
Experimental Method:
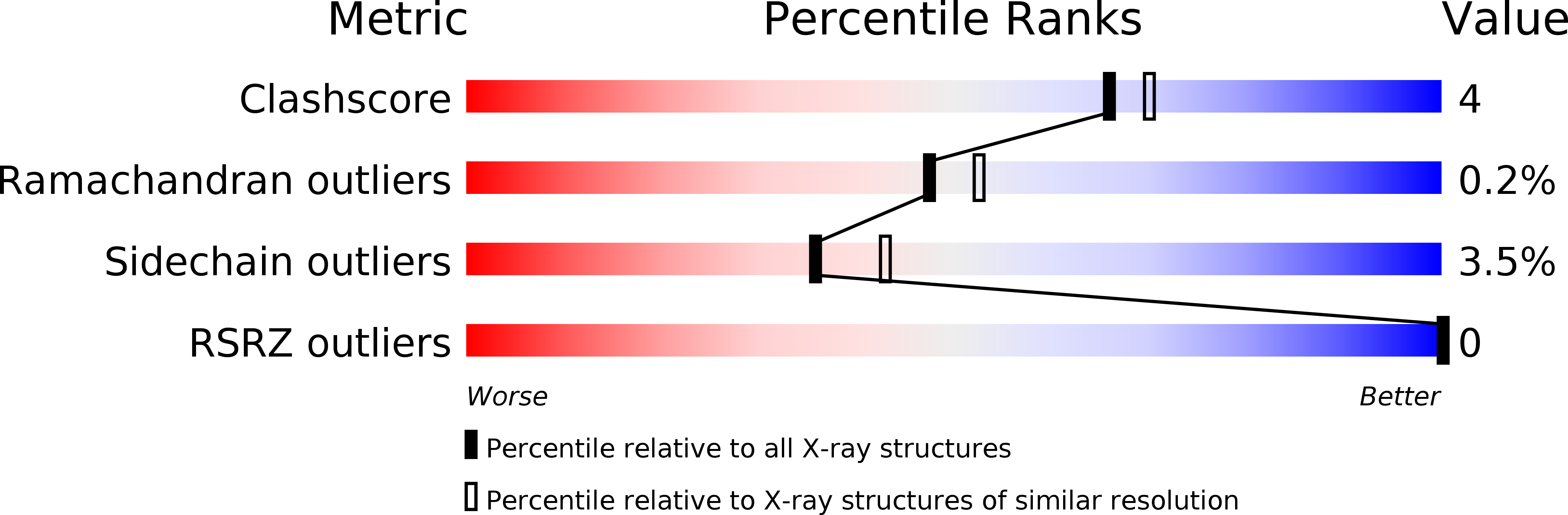
Resolution:
2.18 Å
R-Value Work:
0.13
R-Value Observed:
0.13
Space Group:
C 2 2 21