Deposition Date
2002-04-09
Release Date
2002-04-24
Last Version Date
2024-10-09
Entry Detail
PDB ID:
1LE3
Keywords:
Title:
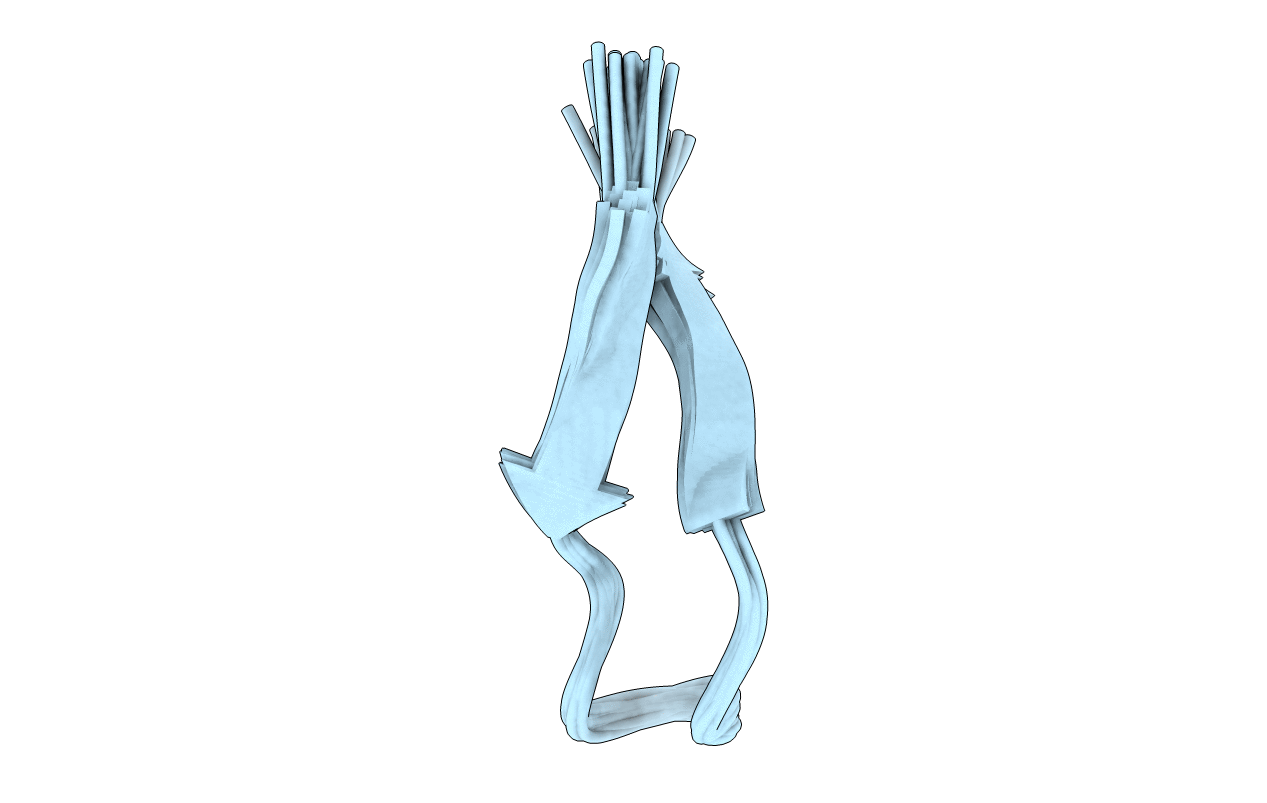
NMR Structure of Tryptophan Zipper 4: A Stable Beta-Hairpin Peptide Based on the C-terminal Hairpin of the B1 Domain of Protein G
Method Details:
Experimental Method:
Conformers Calculated:
64
Conformers Submitted:
20
Selection Criteria:
structures with the least restraint violations