Deposition Date
1995-03-30
Release Date
1995-09-15
Last Version Date
2024-02-14
Entry Detail
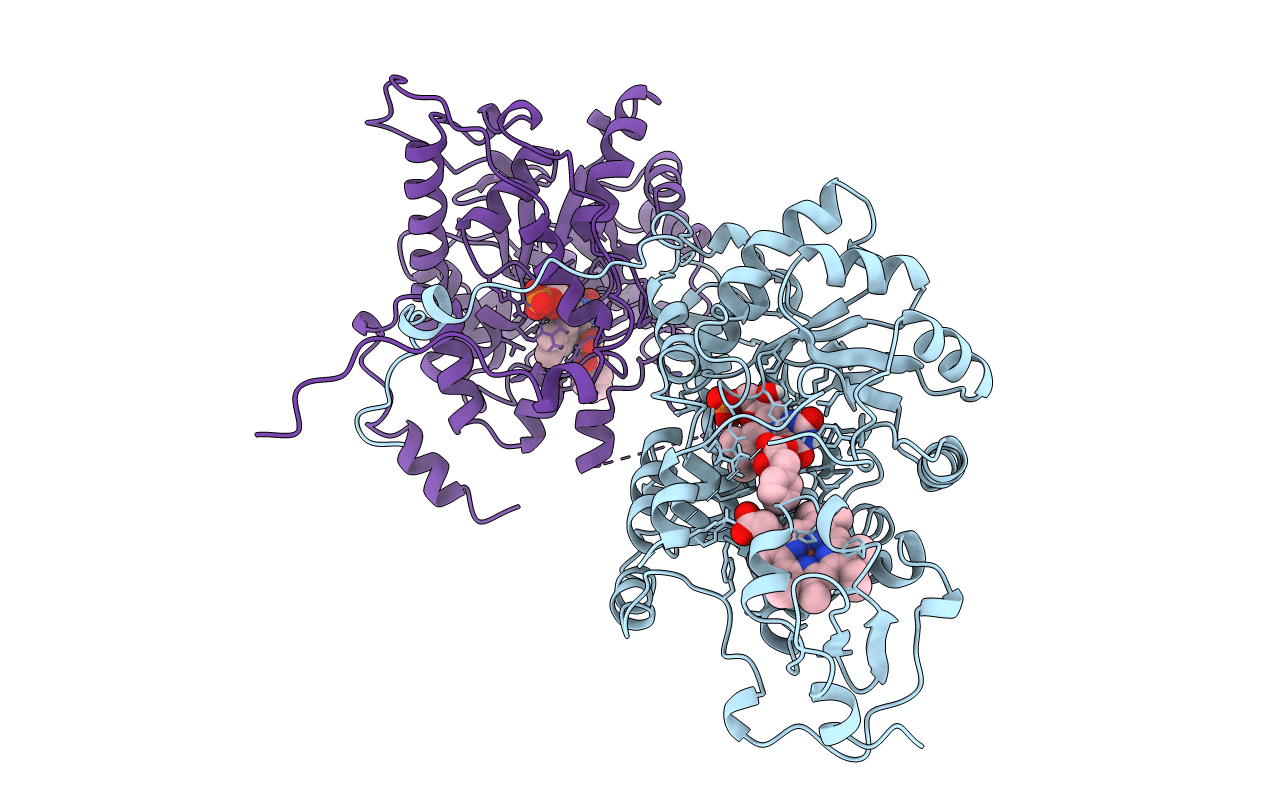
PDB ID:
1LCO
Keywords:
Title:
X-RAY STRUCTURE OF TWO COMPLEXES OF THE Y143F FLAVOCYTOCHROME B2 MUTANT CRYSTALLIZED IN THE PRESENCE OF LACTATE OR PHENYL-LACTATE
Biological Source:
Source Organism(s):
Saccharomyces cerevisiae (Taxon ID: 4932)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.90 Å
R-Value Work:
0.18
R-Value Observed:
0.18
Space Group:
P 32 2 1


