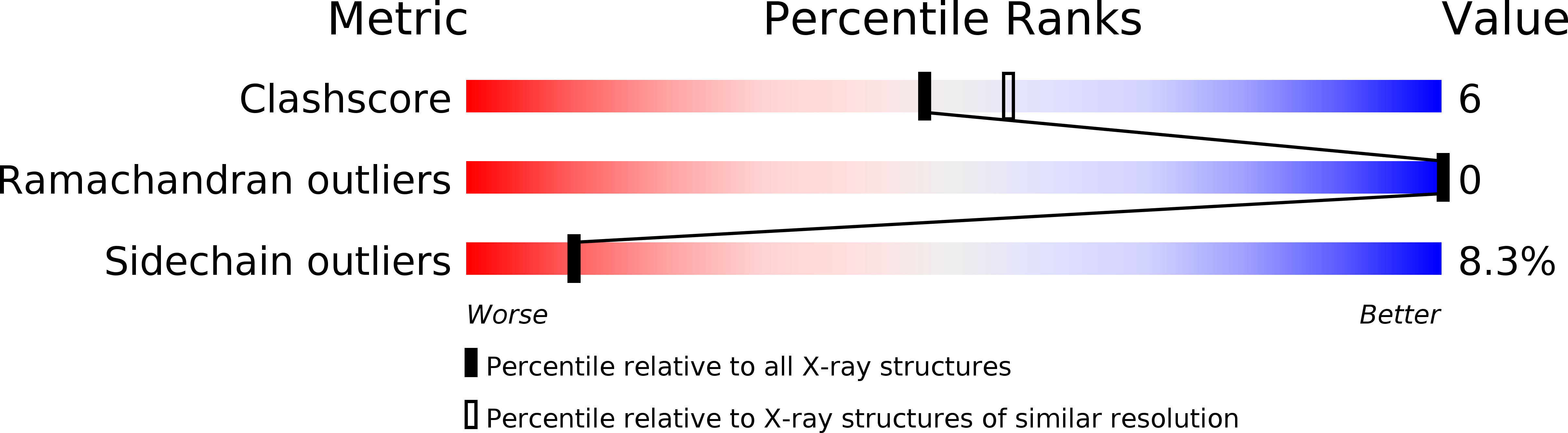Deposition Date
1993-12-22
Release Date
1994-04-30
Last Version Date
2024-02-14
Entry Detail
PDB ID:
1LBA
Keywords:
Title:
THE STRUCTURE OF BACTERIOPHAGE T7 LYSOZYME, A ZINC AMIDASE AND AN INHIBITOR OF T7 RNA POLYMERASE
Biological Source:
Source Organism(s):
Enterobacteria phage T7 (Taxon ID: 10760)
Method Details:
Experimental Method:
Resolution:
2.20 Å
R-Value Work:
0.19
R-Value Observed:
0.19
Space Group:
C 2 2 21