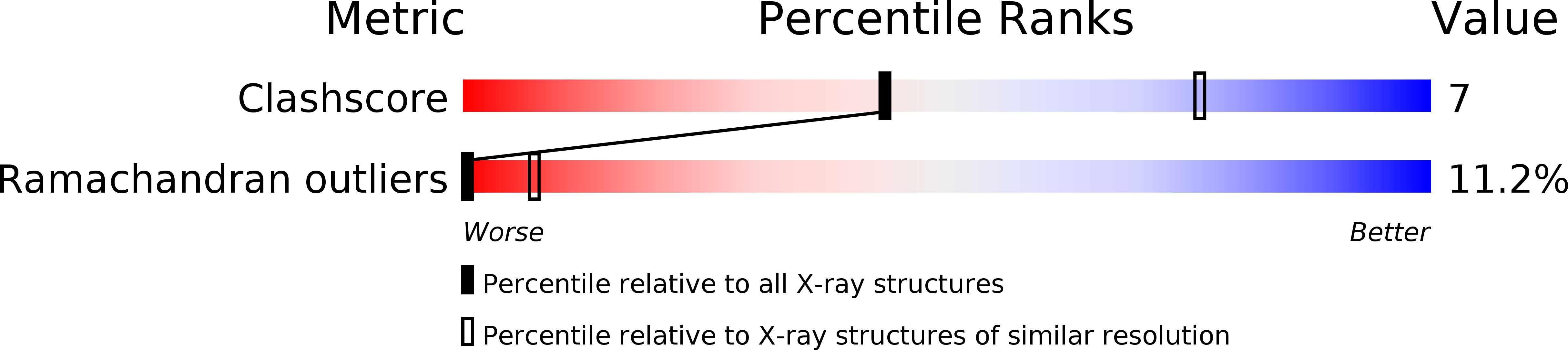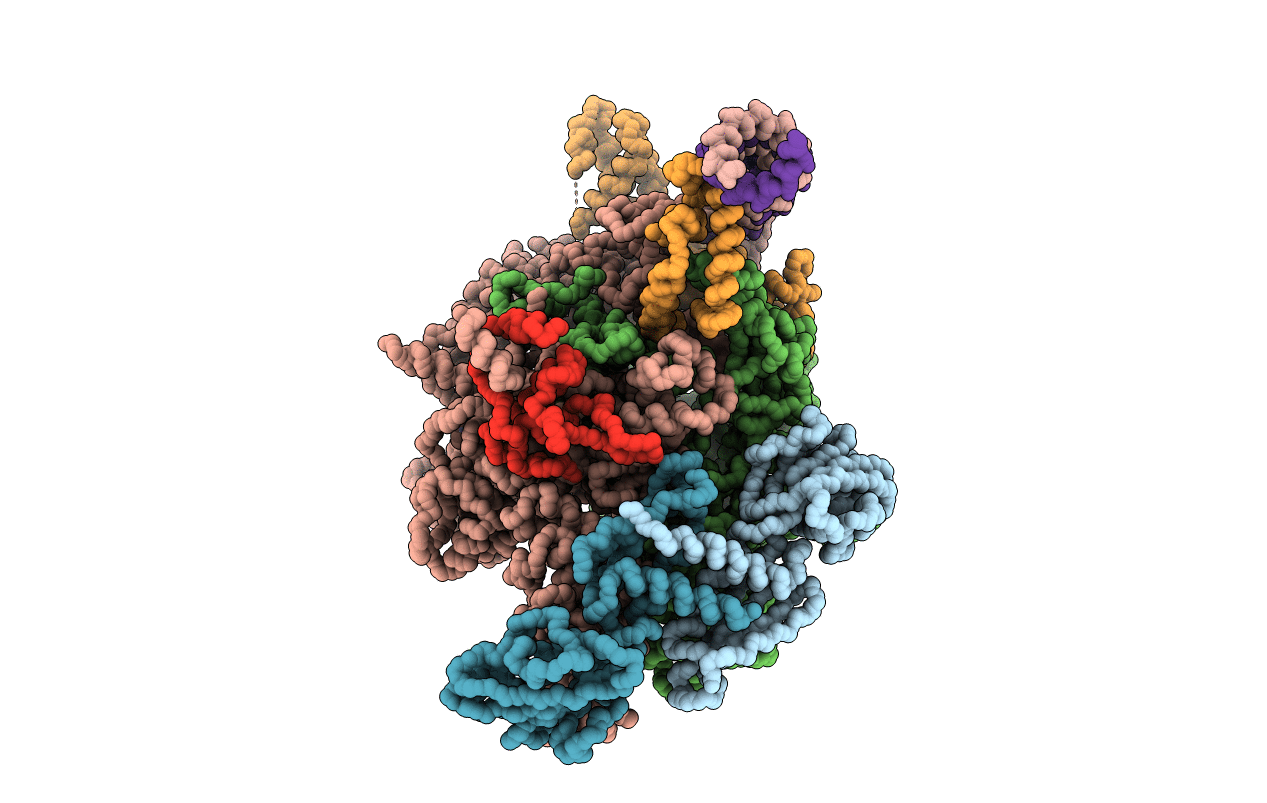
Deposition Date
2002-03-27
Release Date
2002-05-31
Last Version Date
2024-02-14
Entry Detail
PDB ID:
1L9Z
Keywords:
Title:
Thermus aquaticus RNA Polymerase Holoenzyme/Fork-Junction Promoter DNA Complex at 6.5 A Resolution
Biological Source:
Source Organism(s):
Thermus aquaticus (Taxon ID: 271)
Expression System(s):
Method Details: