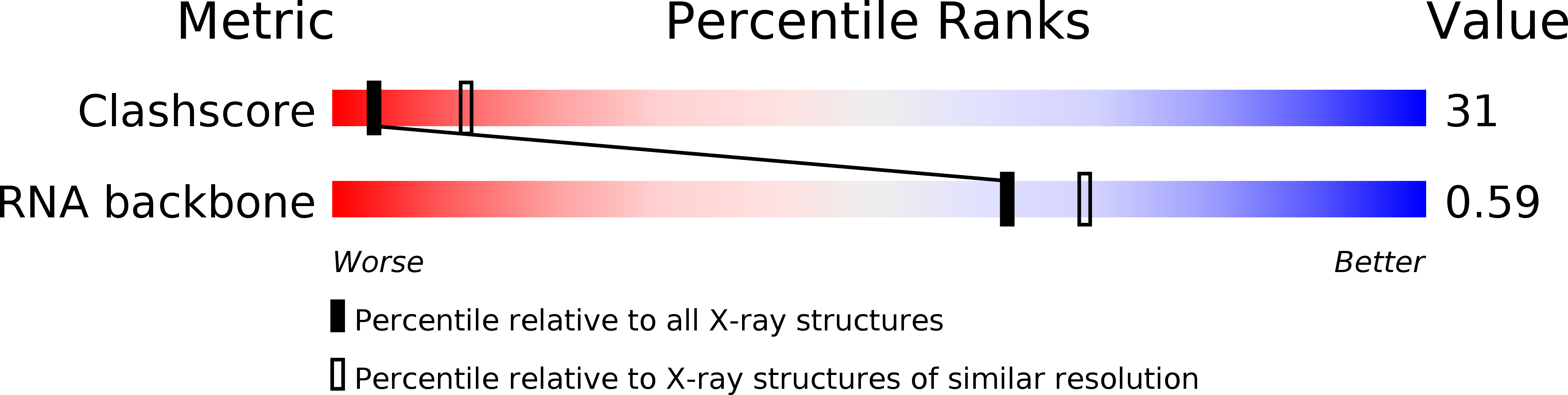
Deposition Date
2002-03-21
Release Date
2002-08-23
Last Version Date
2023-08-16
Entry Detail
PDB ID:
1L8V
Keywords:
Title:
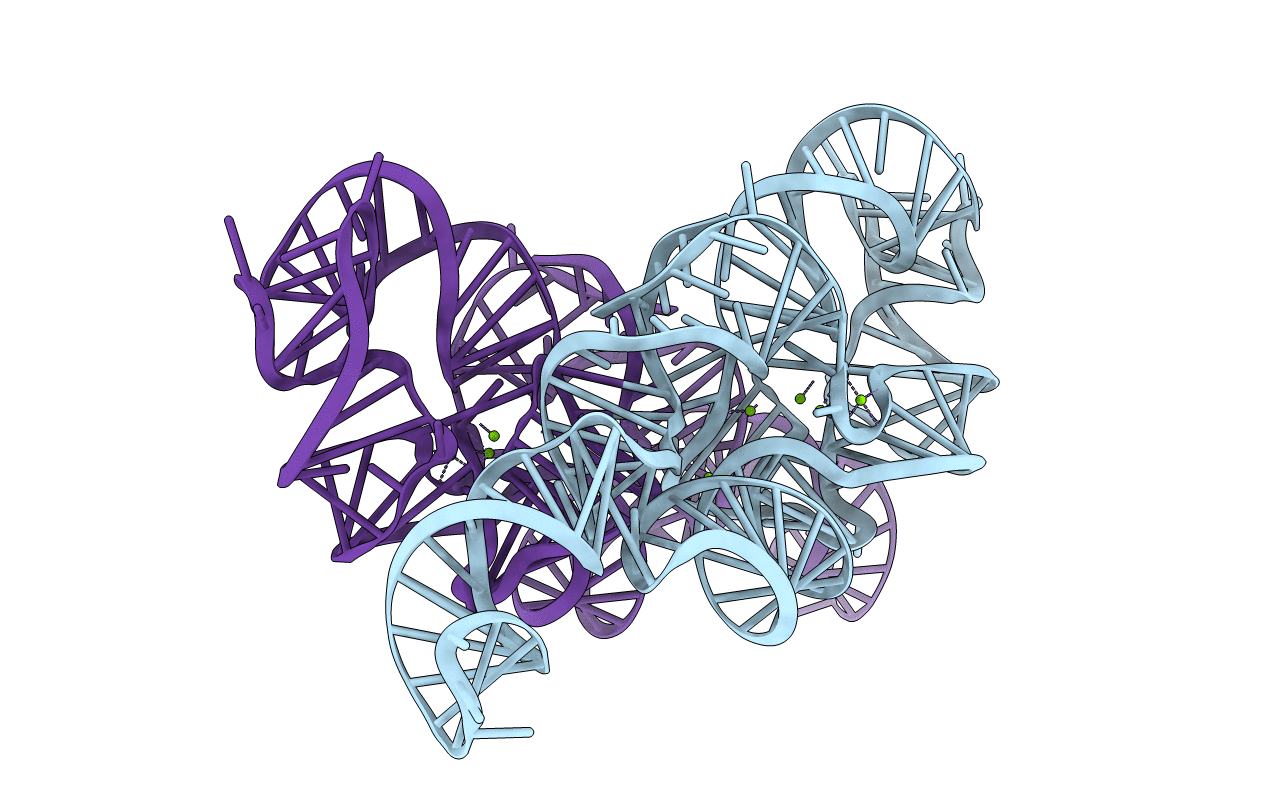
Crystal Structure of a Mutant (C109G,G212C) P4-P6 Domain of the Group I Intron from Tetrahymena Thermophilia
Method Details:
Experimental Method:
Resolution:
2.80 Å
R-Value Free:
0.29
R-Value Work:
0.29
R-Value Observed:
0.29
Space Group:
P 21 21 21