Deposition Date
2002-03-06
Release Date
2002-12-11
Last Version Date
2024-10-16
Entry Detail
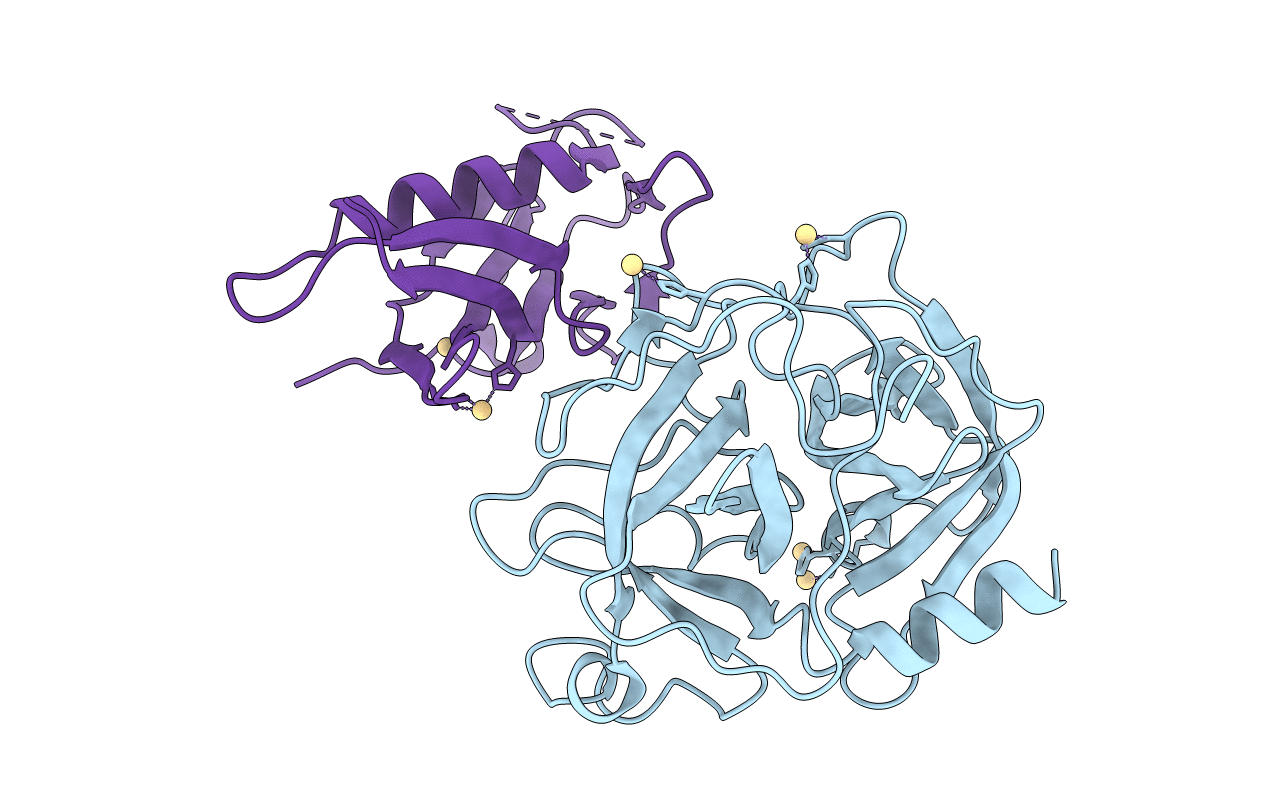
PDB ID:
1L4Z
Keywords:
Title:
X-RAY CRYSTAL STRUCTURE OF THE COMPLEX OF MICROPLASMINOGEN WITH ALPHA DOMAIN OF STREPTOKINASE IN THE PRESENCE CADMIUM IONS
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Streptococcus dysgalactiae subsp. equisimilis (Taxon ID: 119602)
Streptococcus dysgalactiae subsp. equisimilis (Taxon ID: 119602)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.80 Å
R-Value Free:
0.26
R-Value Work:
0.22
Space Group:
P 65 2 2


