Deposition Date
2001-12-19
Release Date
2002-01-17
Last Version Date
2024-02-14
Entry Detail
PDB ID:
1KNZ
Keywords:
Title:
Recognition of the rotavirus mRNA 3' consensus by an asymmetric NSP3 homodimer
Biological Source:
Source Organism:
Simian rotavirus A/SA11 (Taxon ID: 10923)
Host Organism:
Method Details:
Experimental Method:
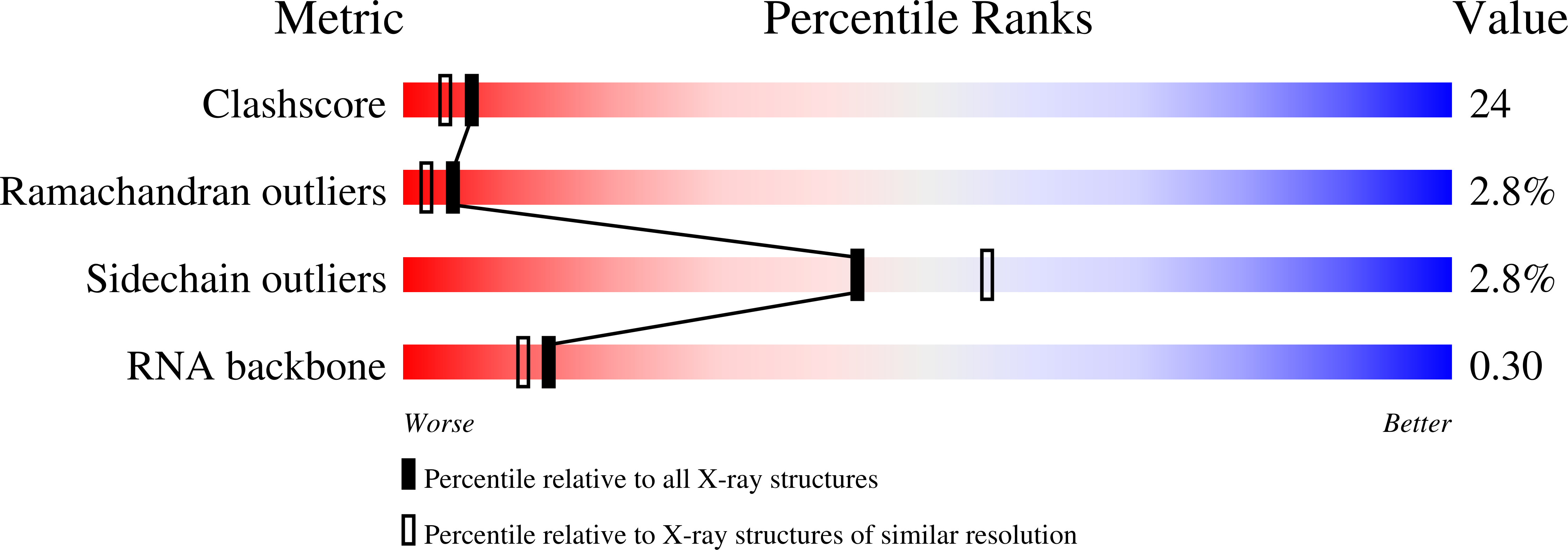
Resolution:
2.45 Å
R-Value Free:
0.28
R-Value Work:
0.22
Space Group:
P 1