Deposition Date
2001-11-21
Release Date
2002-08-23
Last Version Date
2024-11-13
Entry Detail
PDB ID:
1KFL
Keywords:
Title:
Crystal structure of phenylalanine-regulated 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase (DAHP synthase) from E.coli complexed with Mn2+, PEP, and Phe
Biological Source:
Source Organism(s):
Escherichia coli (Taxon ID: 562)
Expression System(s):
Method Details:
Experimental Method:
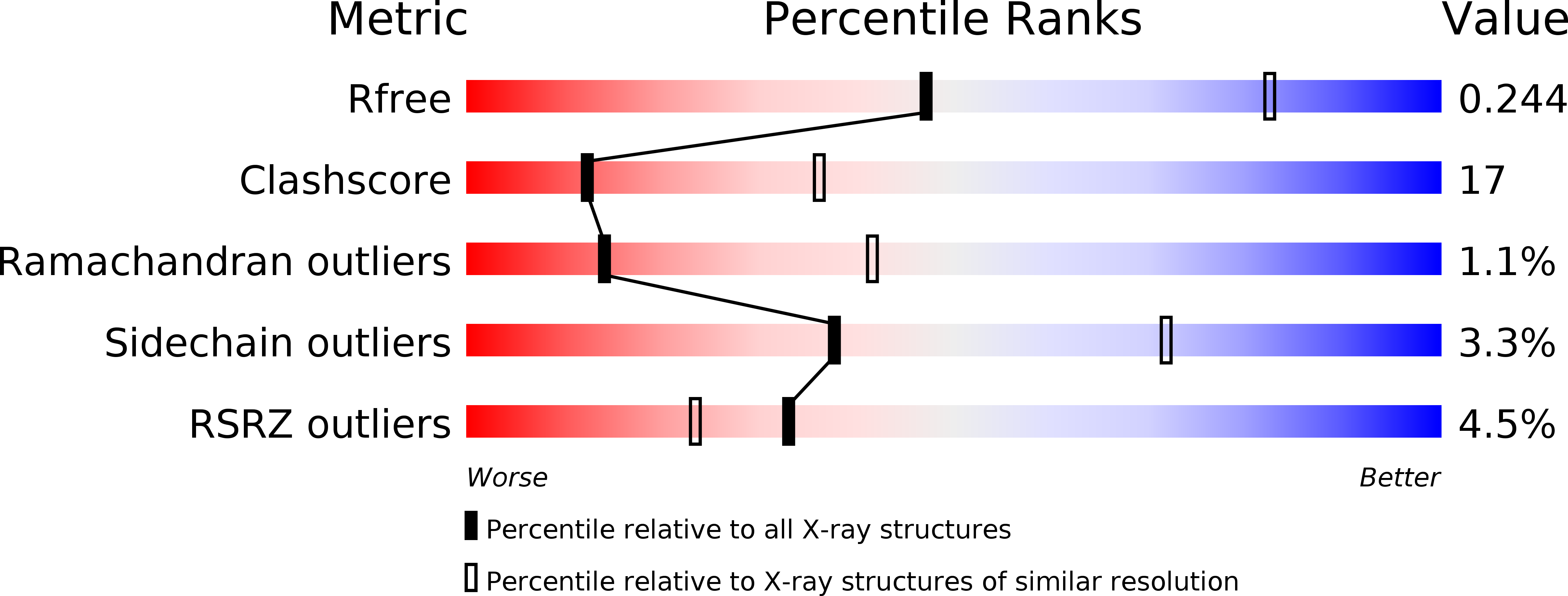
Resolution:
2.80 Å
R-Value Free:
0.24
R-Value Work:
0.21
R-Value Observed:
0.21
Space Group:
C 1 2 1