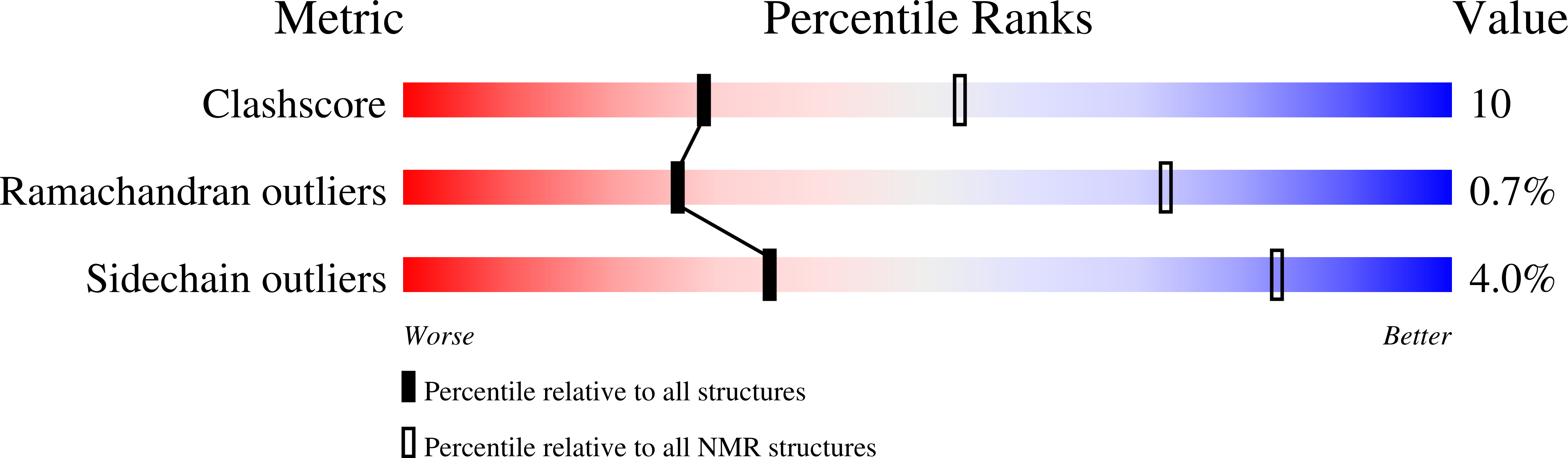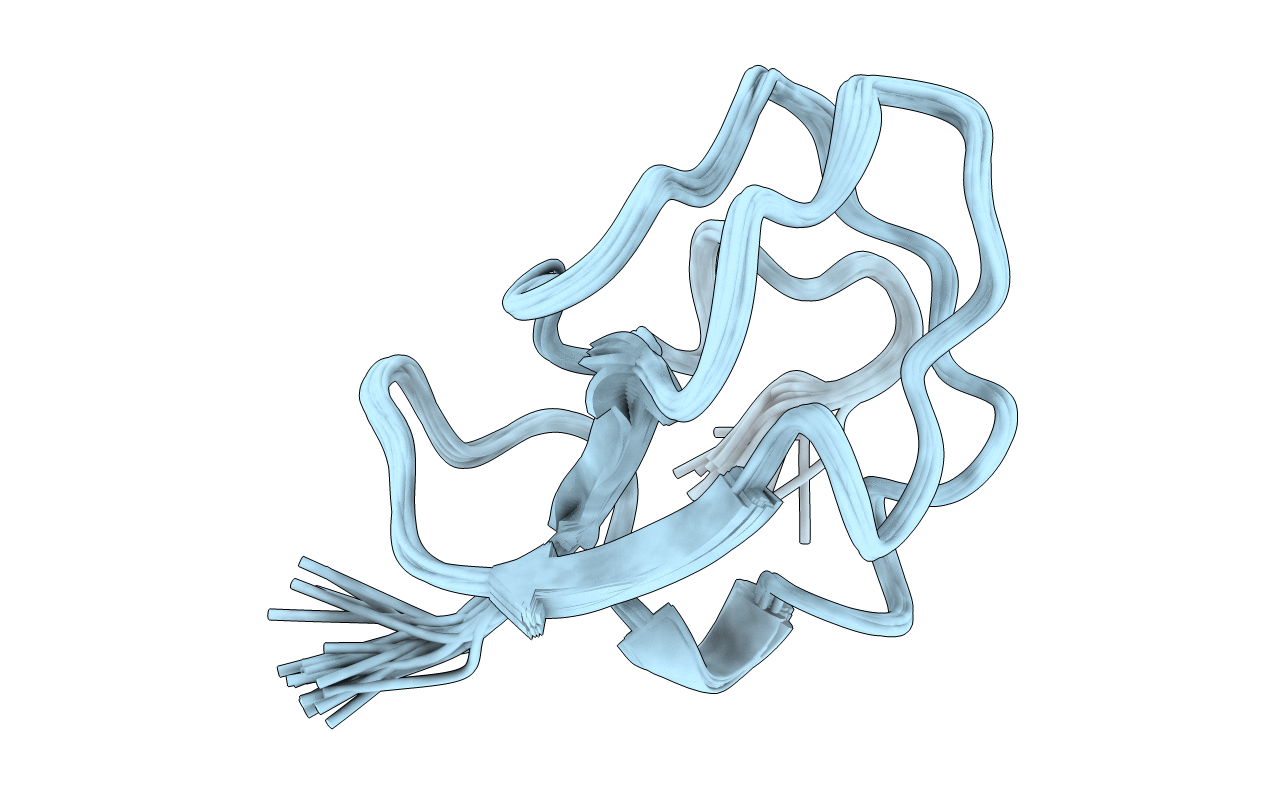
Deposition Date
1996-07-08
Release Date
1997-04-21
Last Version Date
2024-05-22
Entry Detail
PDB ID:
1KDE
Keywords:
Title:
NORTH-ATLANTIC OCEAN POUT ANTIFREEZE PROTEIN TYPE III ISOFORM HPLC12 MUTANT, NMR, 22 STRUCTURES
Biological Source:
Source Organism(s):
Macrozoarces americanus (Taxon ID: 8199)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
50
Conformers Submitted:
22
Selection Criteria:
X-PLOR ENERGIES