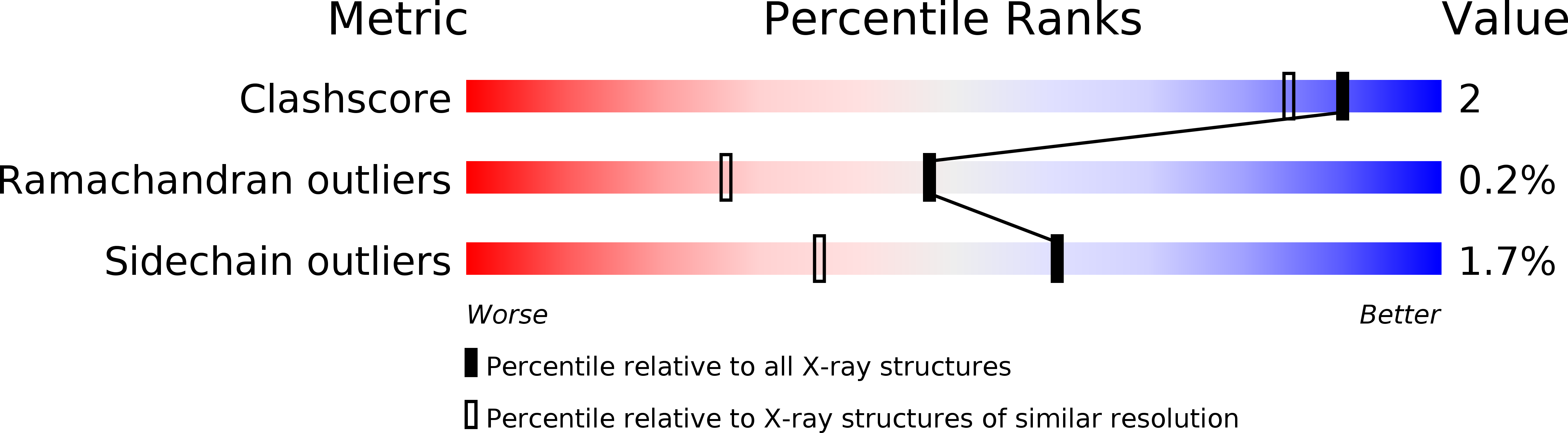
Deposition Date
1995-06-08
Release Date
1995-10-15
Last Version Date
2024-02-07
Entry Detail
PDB ID:
1KAP
Keywords:
Title:
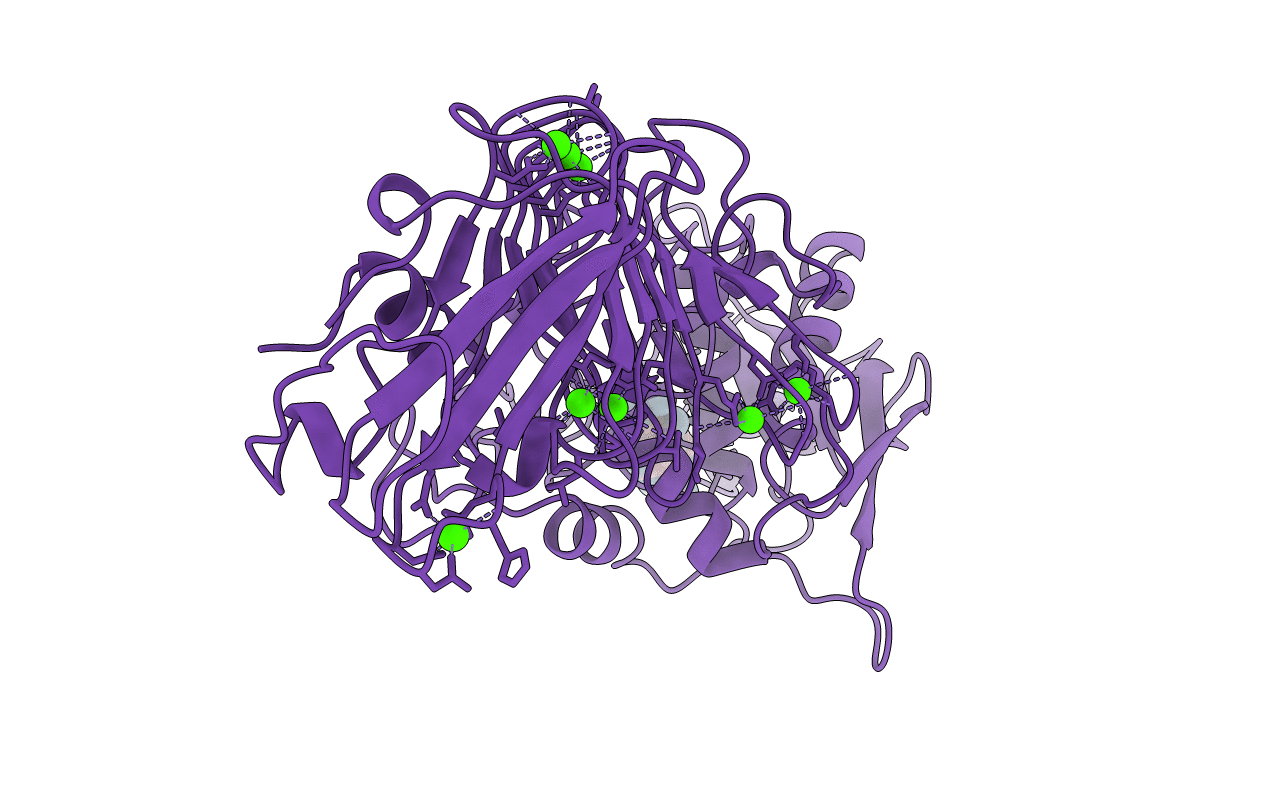
THREE-DIMENSIONAL STRUCTURE OF THE ALKALINE PROTEASE OF PSEUDOMONAS AERUGINOSA: A TWO-DOMAIN PROTEIN WITH A CALCIUM BINDING PARALLEL BETA ROLL MOTIF
Biological Source:
Source Organism(s):
Pseudomonas aeruginosa (Taxon ID: 208964)
Method Details:
Experimental Method:
Resolution:
1.64 Å
R-Value Work:
0.17
R-Value Observed:
0.17
Space Group:
P 65