Deposition Date
2001-10-26
Release Date
2002-06-19
Last Version Date
2023-08-16
Entry Detail
PDB ID:
1K8Y
Keywords:
Title:
CRYSTAL STRUCTURE OF THE TRYPTOPHAN SYNTHASE BETA-SER178PRO MUTANT COMPLEXED WITH D,L-ALPHA-GLYCEROL-3-PHOSPHATE
Biological Source:
Source Organism(s):
Salmonella typhimurium (Taxon ID: 602)
Expression System(s):
Method Details:
Experimental Method:
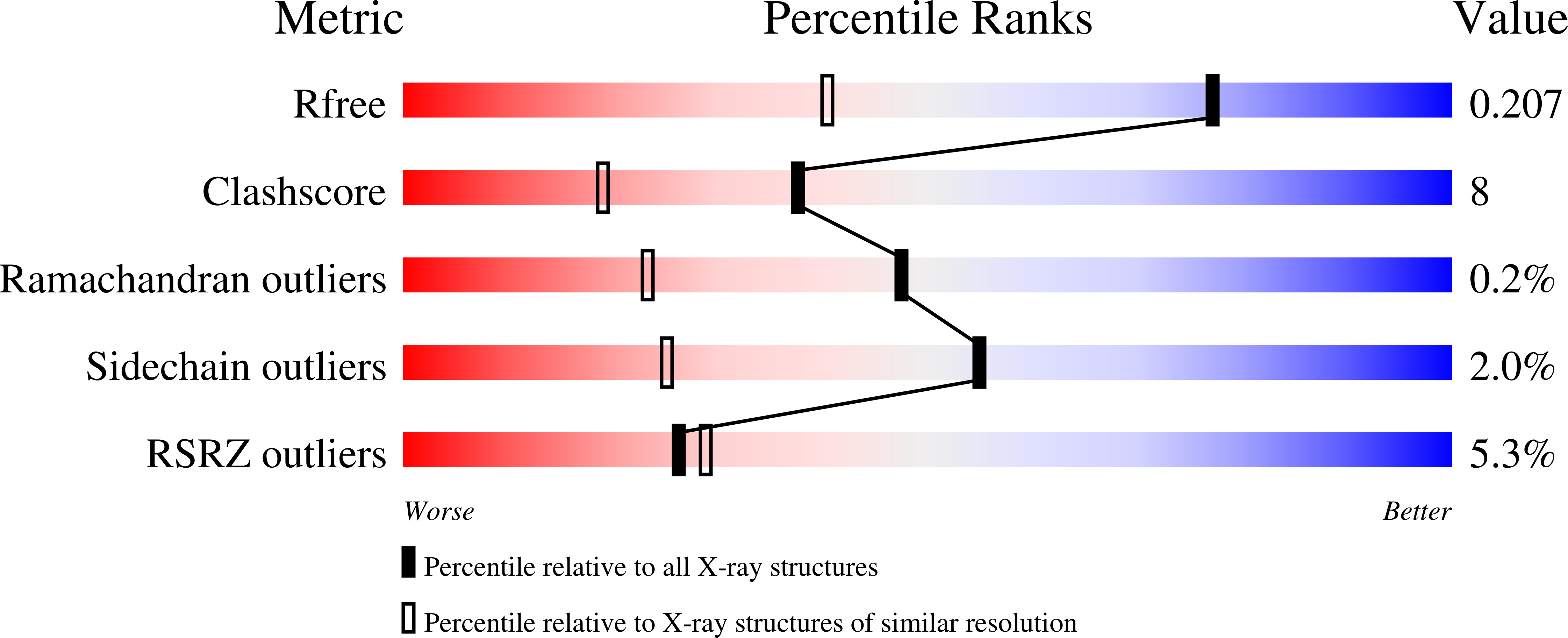
Resolution:
1.50 Å
R-Value Free:
0.20
R-Value Work:
0.17
R-Value Observed:
0.17
Space Group:
C 1 2 1