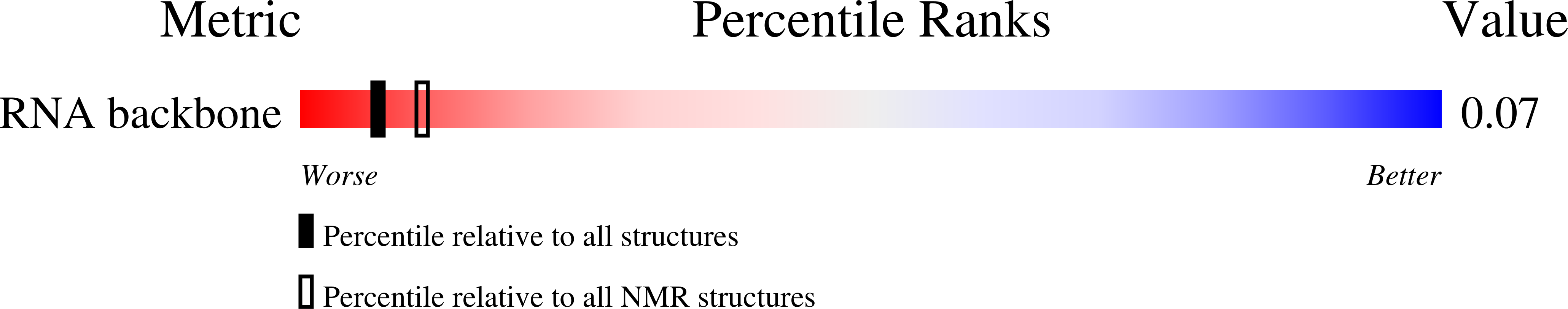
Deposition Date
2001-09-03
Release Date
2002-03-20
Last Version Date
2024-05-22
Entry Detail
PDB ID:
1JWC
Keywords:
Title:
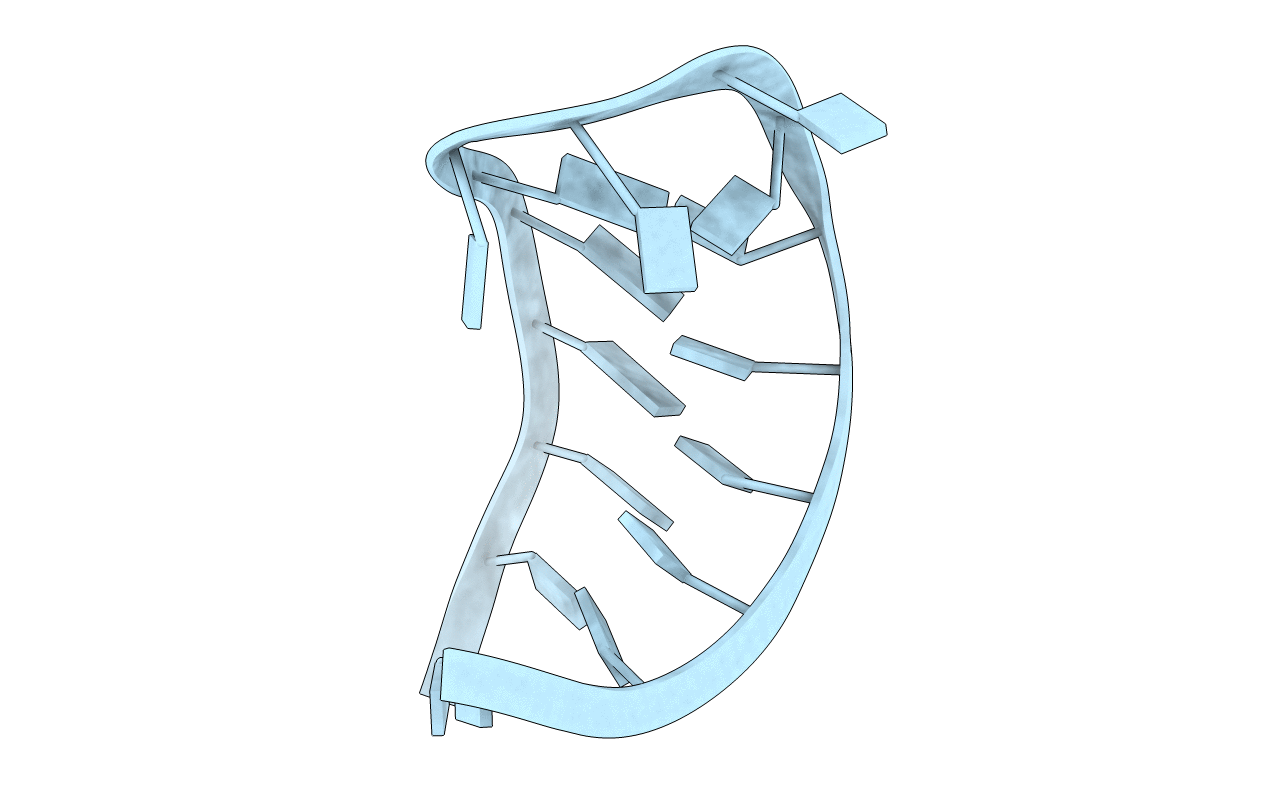
NMR SOLUTION STRUCTURE OF THE RNA HAIRPIN BINDING SITE FOR THE HISTONE STEM-LOOP BINDING PROTEIN
Method Details:
Experimental Method:
Conformers Submitted:
1