Deposition Date
1995-12-07
Release Date
1996-03-08
Last Version Date
2024-11-20
Entry Detail
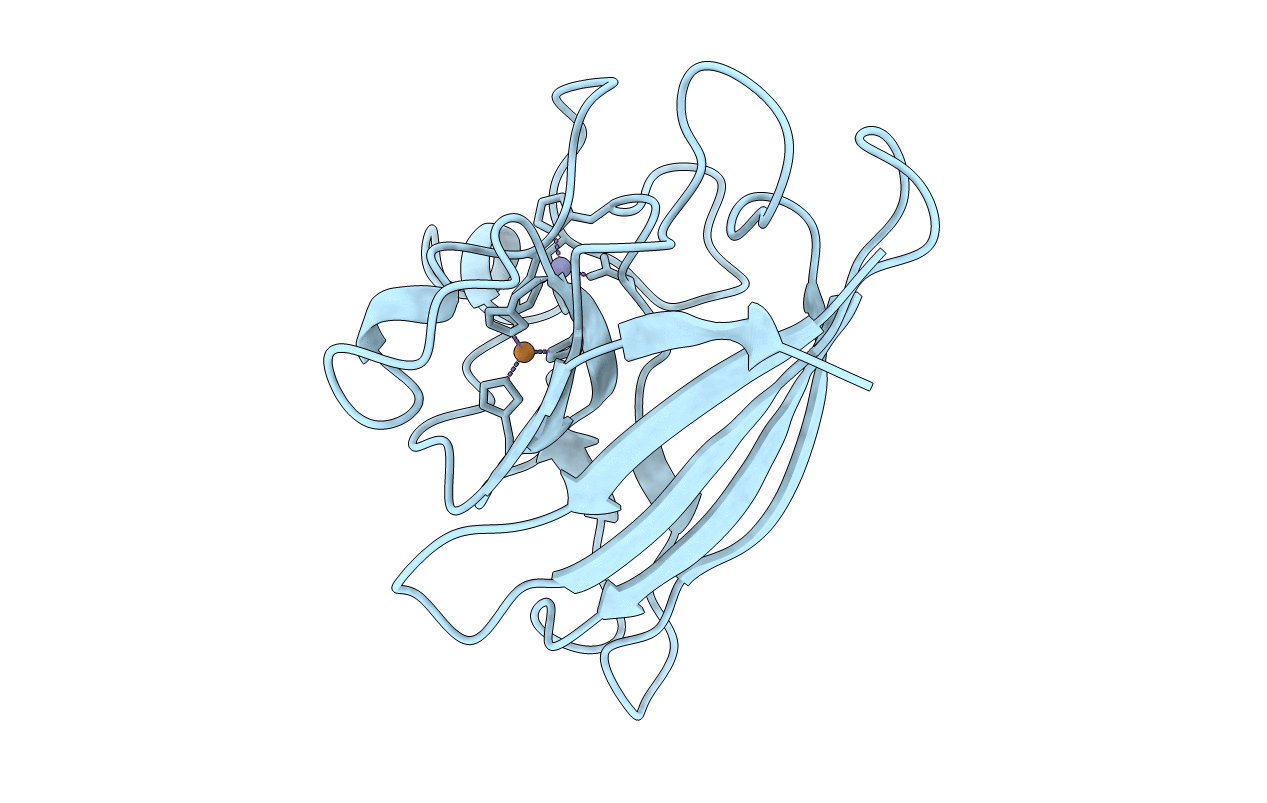
PDB ID:
1JCV
Keywords:
Title:
REDUCED BRIDGE-BROKEN YEAST CU/ZN SUPEROXIDE DISMUTASE LOW TEMPERATURE (-180C) STRUCTURE
Biological Source:
Source Organism(s):
Saccharomyces cerevisiae (Taxon ID: 4932)
Method Details:


