Deposition Date
2001-05-21
Release Date
2002-02-27
Last Version Date
2024-11-13
Entry Detail
PDB ID:
1J8D
Keywords:
Title:
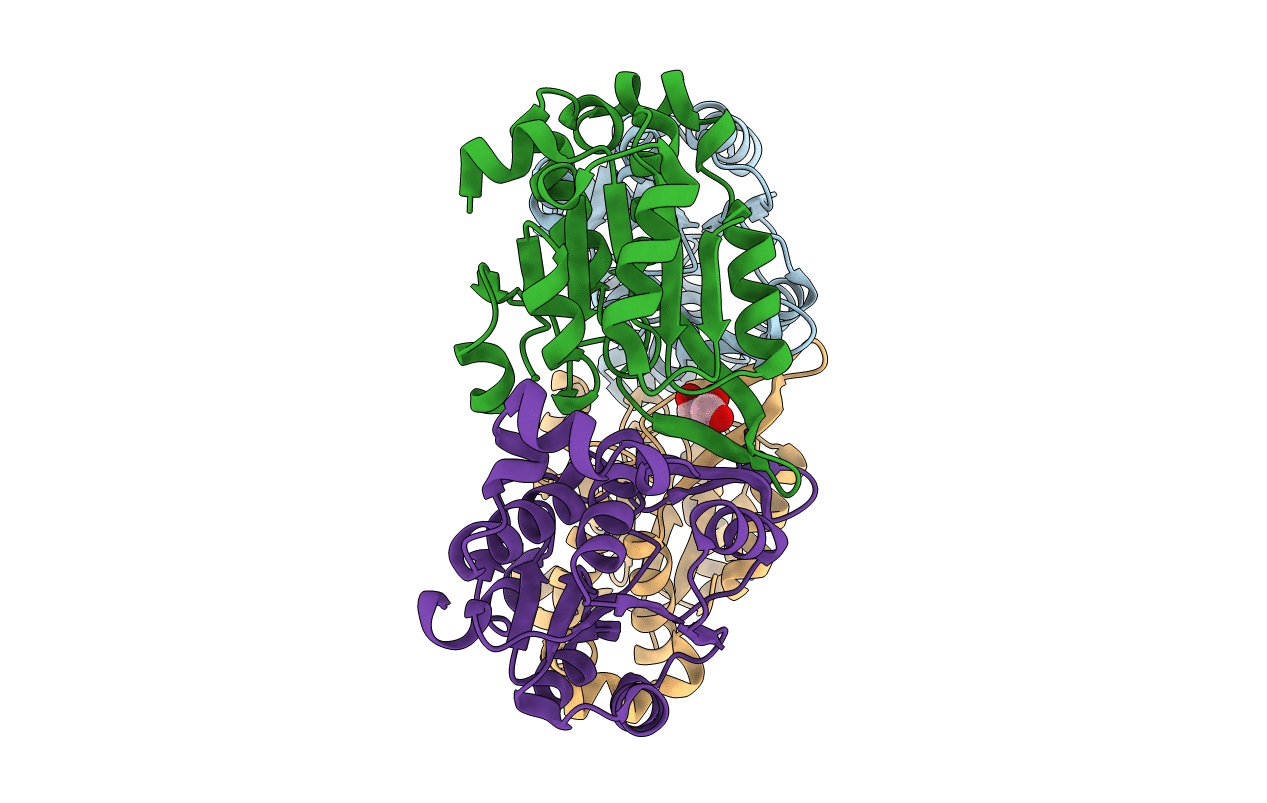
Structure Of the metal-free form of the deoxy-D-mannose-octulosonate 8-phosphate phosphatase (YrbI) From Haemophilus Influenzae (HI1679)
Biological Source:
Source Organism(s):
Haemophilus influenzae Rd (Taxon ID: 71421)
Expression System(s):
Method Details:
Experimental Method:
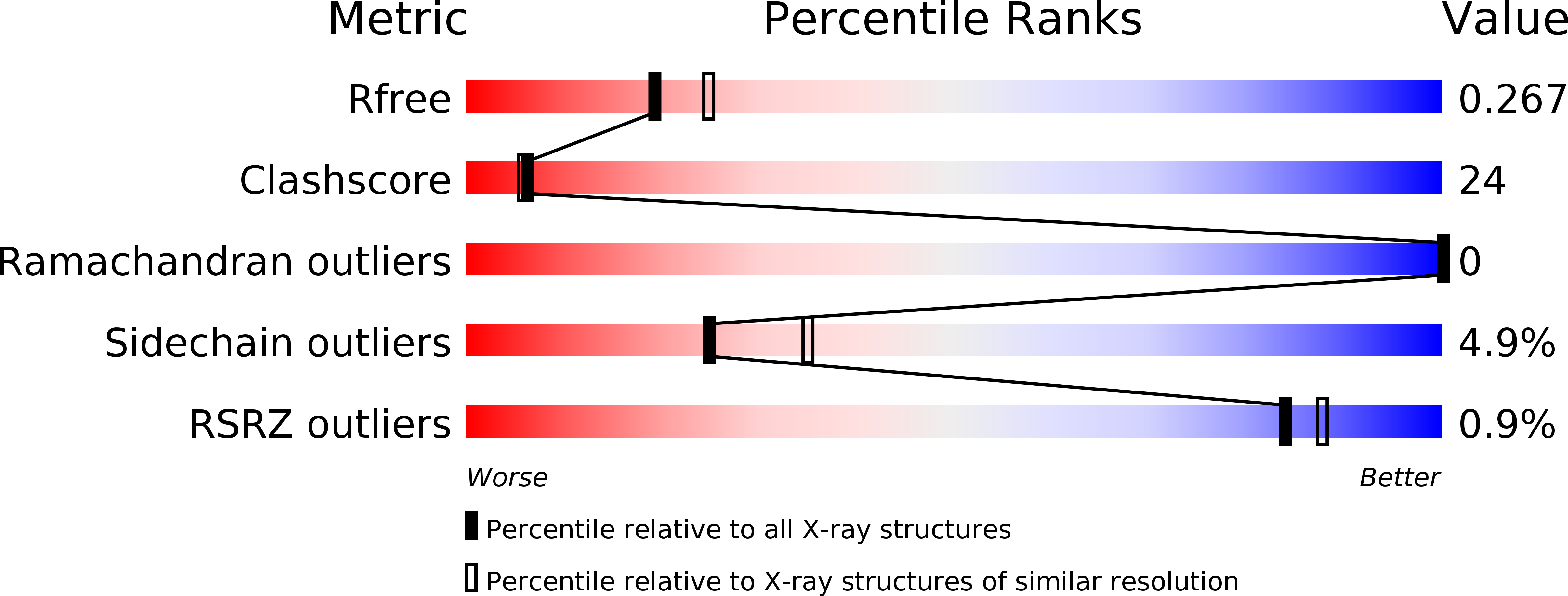
Resolution:
2.30 Å
R-Value Free:
0.25
R-Value Work:
0.17
Space Group:
I 2 2 2