Deposition Date
2001-08-18
Release Date
2002-05-29
Last Version Date
2024-04-03
Entry Detail
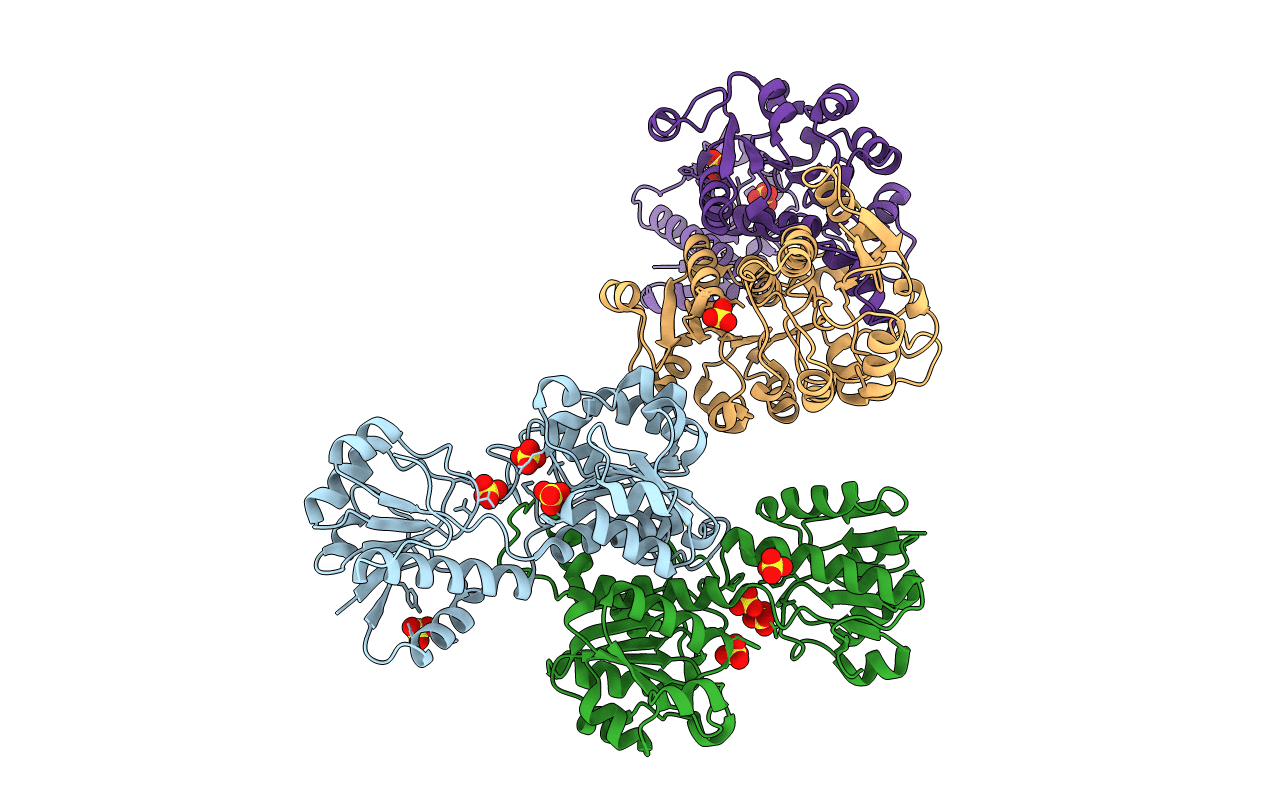
PDB ID:
1J4A
Keywords:
Title:
INSIGHTS INTO DOMAIN CLOSURE, SUBSTRATE SPECIFICITY AND CATALYSIS OF D-LACTATE DEHYDROGENASE FROM LACTOBACILLUS BULGARICUS
Biological Source:
Source Organism:
Lactobacillus delbrueckii subsp. bulgaricus (Taxon ID: 1585)
Host Organism:
Method Details:
Experimental Method:
Resolution:
1.90 Å
R-Value Free:
0.25
R-Value Work:
0.20
R-Value Observed:
0.20
Space Group:
C 2 2 21


