Deposition Date
2001-08-31
Release Date
2002-03-13
Last Version Date
2023-11-15
Entry Detail
PDB ID:
1IR1
Keywords:
Title:
Crystal Structure of Spinach Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase (Rubisco) Complexed with CO2, Mg2+ and 2-Carboxyarabinitol-1,5-Bisphosphate
Biological Source:
Source Organism(s):
Spinacia oleracea (Taxon ID: 3562)
Method Details:
Experimental Method:
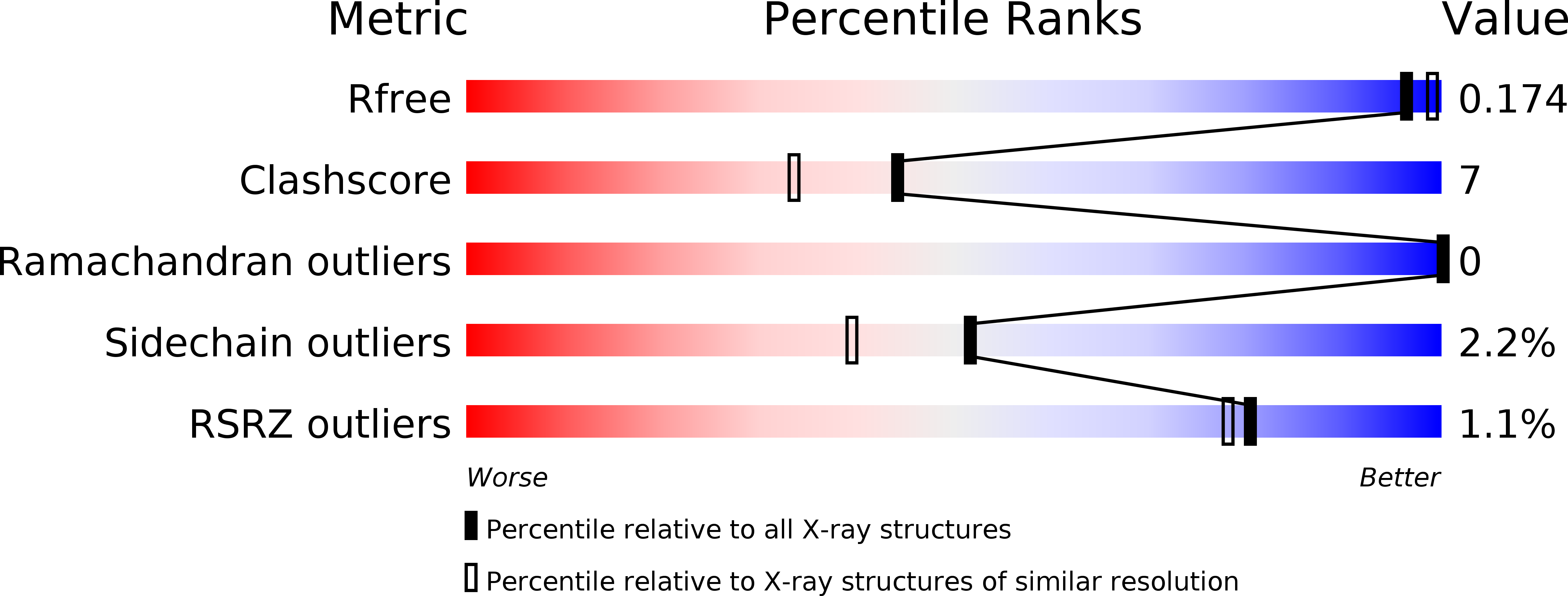
Resolution:
1.80 Å
R-Value Free:
0.17
R-Value Work:
0.15
Space Group:
C 2 2 21