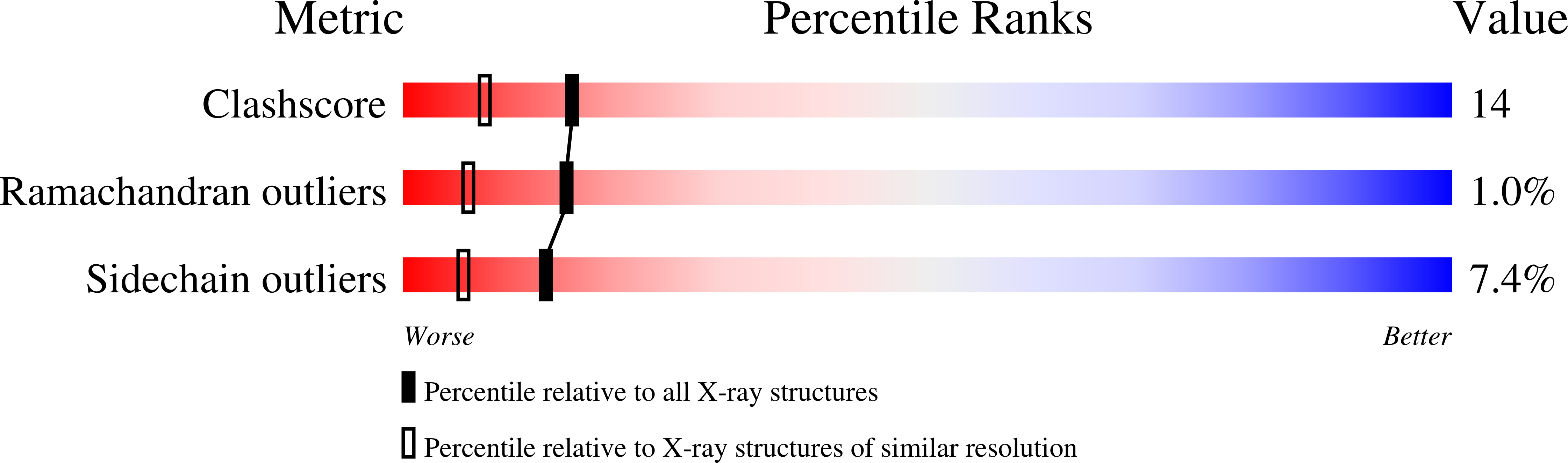
Deposition Date
1996-09-20
Release Date
1997-02-12
Last Version Date
2024-05-22
Entry Detail
PDB ID:
1IOW
Keywords:
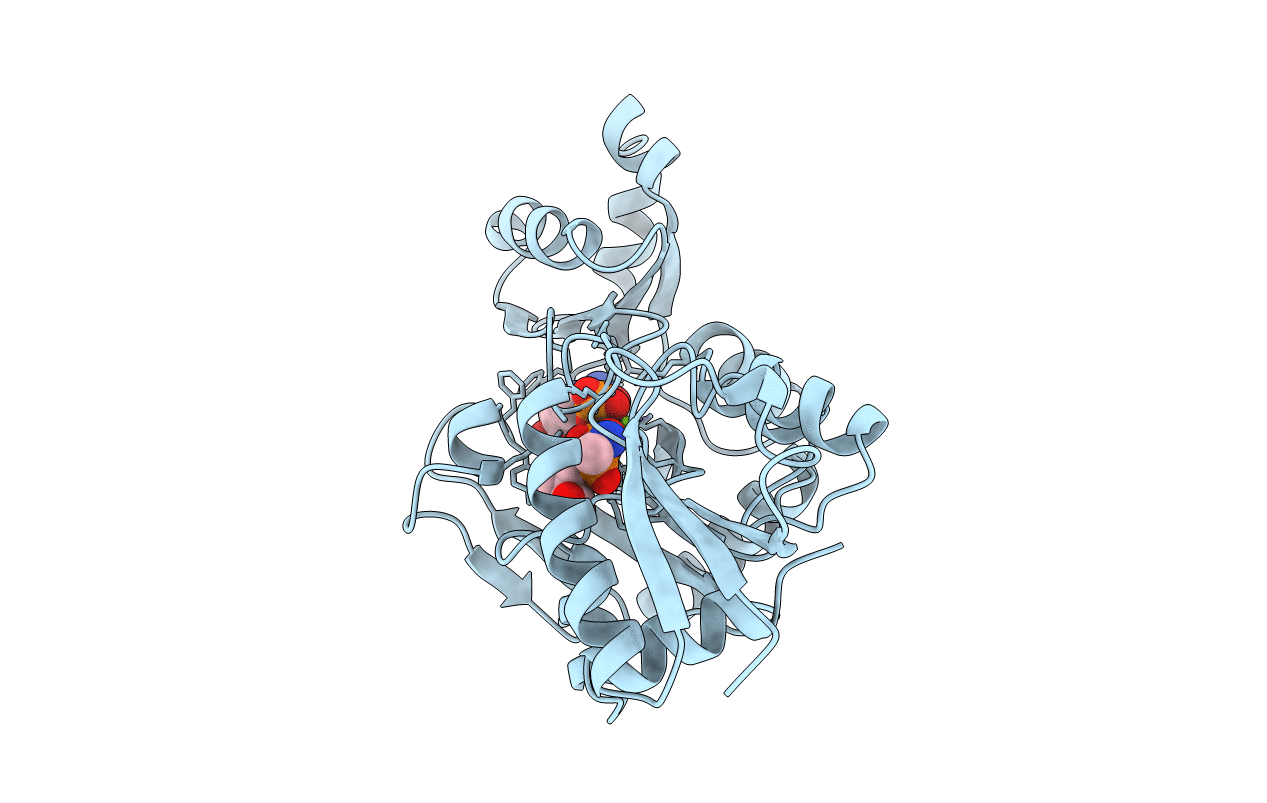
Title:
COMPLEX OF Y216F D-ALA:D-ALA LIGASE WITH ADP AND A PHOSPHORYL PHOSPHINATE
Biological Source:
Source Organism(s):
Escherichia coli (Taxon ID: 562)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
1.90 Å
R-Value Free:
0.23
R-Value Work:
0.15
R-Value Observed:
0.15
Space Group:
P 21 21 2