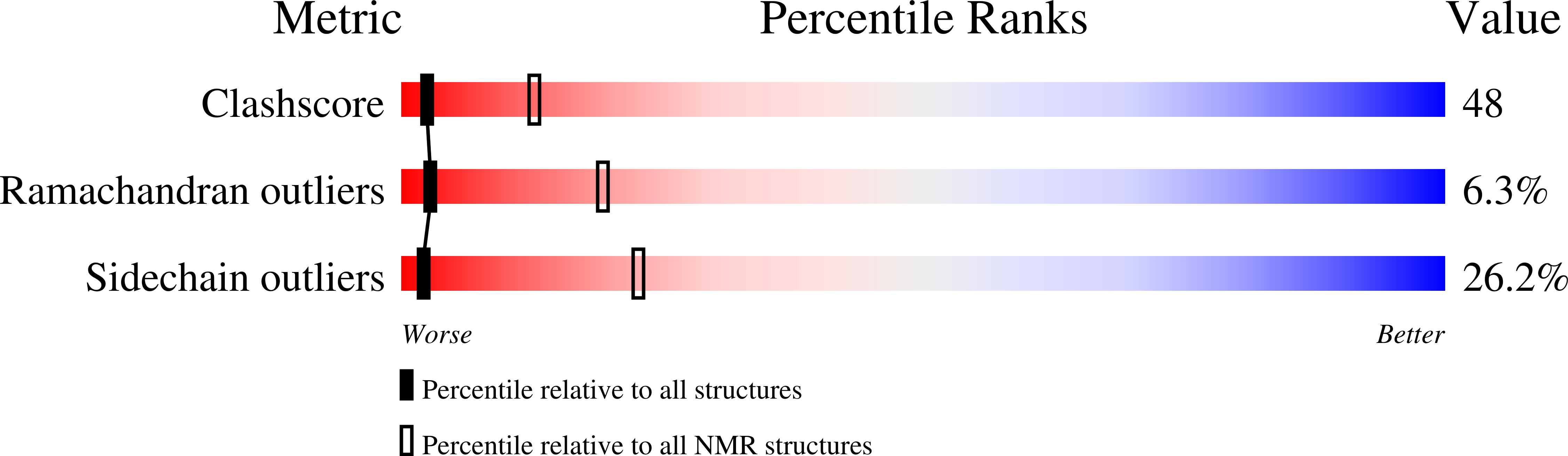Deposition Date
1998-08-13
Release Date
1999-01-13
Last Version Date
2024-10-30
Entry Detail
PDB ID:
1IOH
Keywords:
Title:
INSULIN MUTANT A8 HIS,(B1, B10, B16, B27)GLU, DES-B30, NMR, 26 STRUCTURES
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Expression System(s):
Method Details: