Deposition Date
2001-04-24
Release Date
2001-10-24
Last Version Date
2024-11-13
Entry Detail
PDB ID:
1IIY
Keywords:
Title:
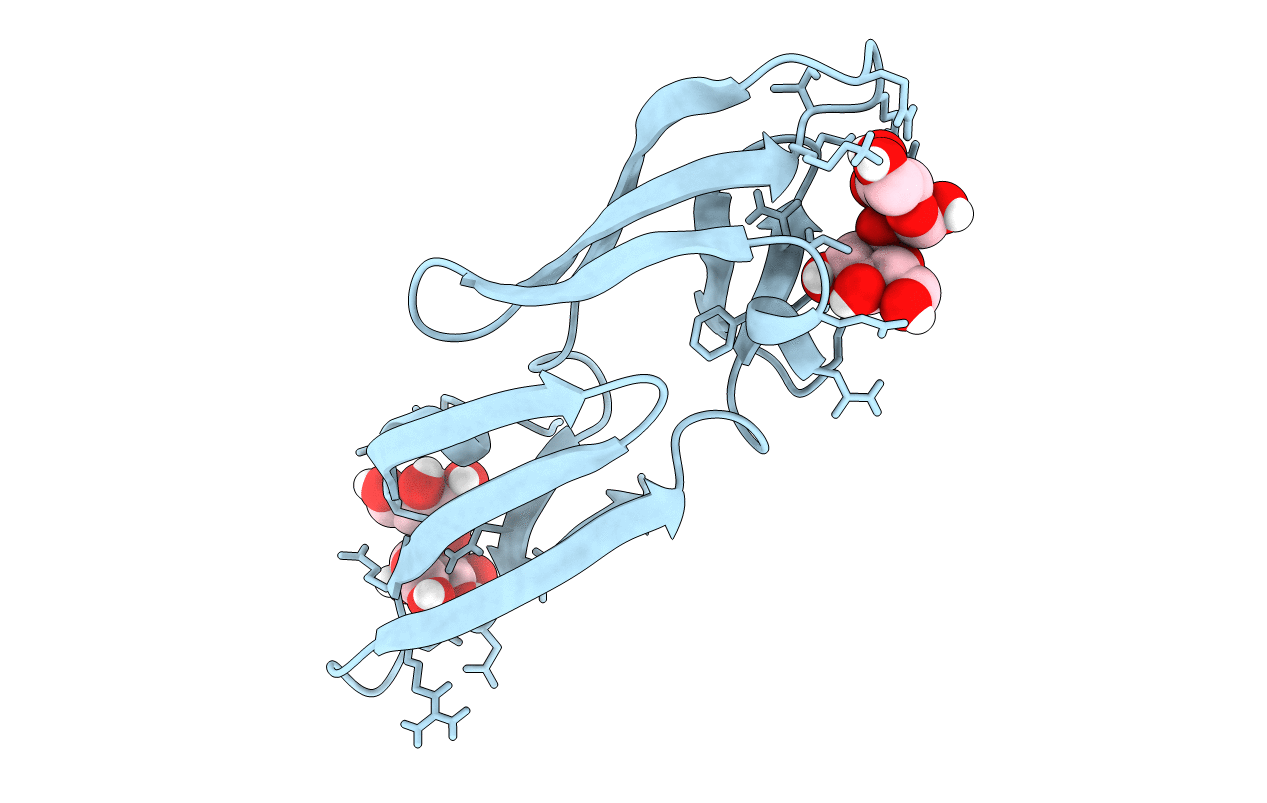
Solution NMR Structure of Complex of 1:2 Cyanovirin-N:Man-Alpha1,2-Man-Alpha Restrained Regularized Mean Coordinates
Biological Source:
Source Organism(s):
Nostoc ellipsosporum (Taxon ID: 45916)
Method Details: