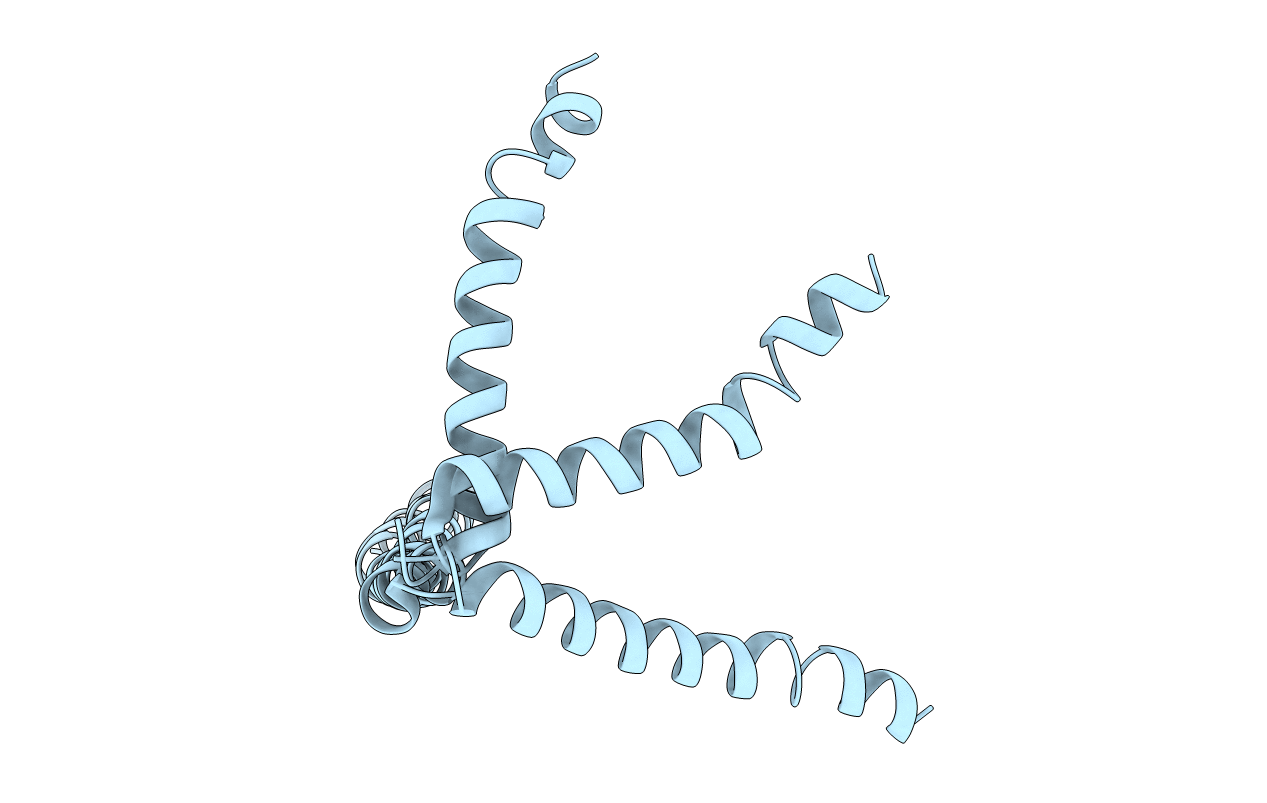
Deposition Date
2001-04-23
Release Date
2001-06-27
Last Version Date
2024-05-22
Entry Detail
PDB ID:
1IIJ
Keywords:
Title:
SOLUTION STRUCTURE OF THE NEU/ERBB-2 MEMBRANE SPANNING SEGMENT
Method Details:
Experimental Method:
Conformers Calculated:
20
Conformers Submitted:
5
Selection Criteria:
structures with the lowest energy