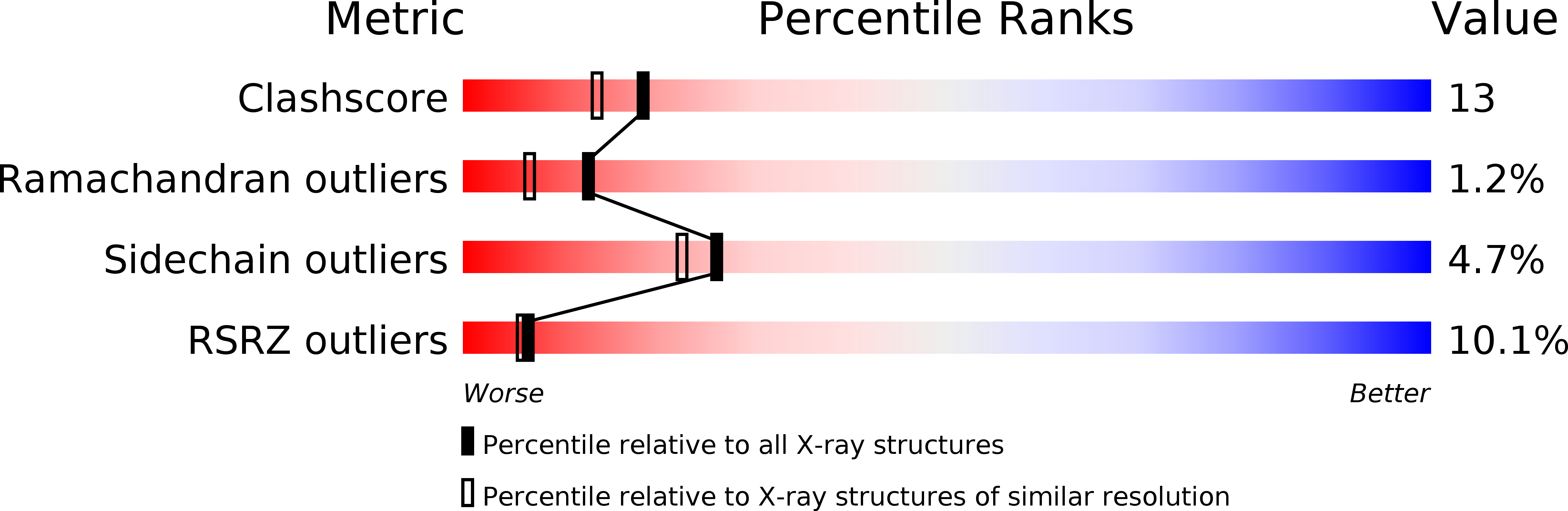
Deposition Date
2001-03-21
Release Date
2001-05-16
Last Version Date
2024-02-07
Entry Detail
PDB ID:
1I9Y
Keywords:
Title:
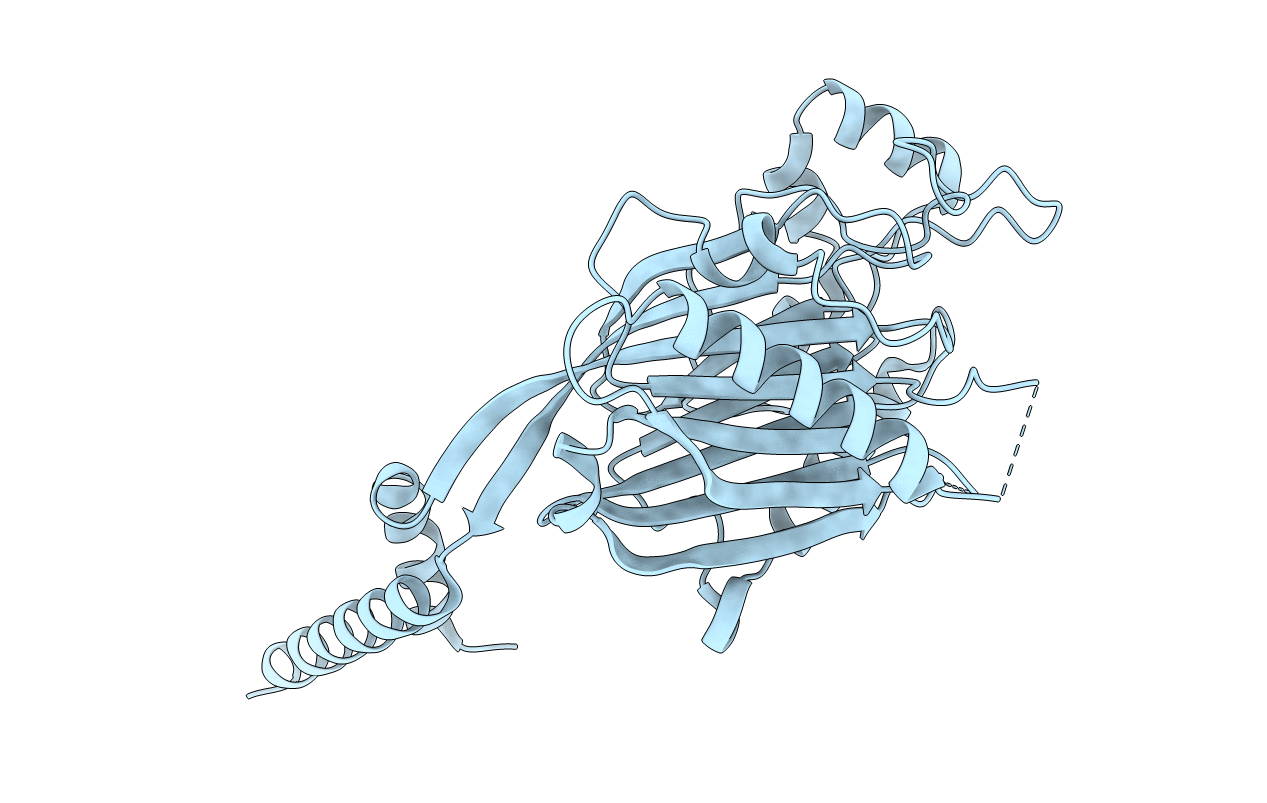
CRYSTAL STRUCTURE OF INOSITOL POLYPHOSPHATE 5-PHOSPHATASE DOMAIN (IPP5C) OF SPSYNAPTOJANIN
Biological Source:
Source Organism:
Schizosaccharomyces pombe (Taxon ID: 4896)
Host Organism:
Method Details:
Experimental Method:
Resolution:
2.00 Å
R-Value Free:
0.22
R-Value Work:
0.19
Space Group:
C 1 2 1