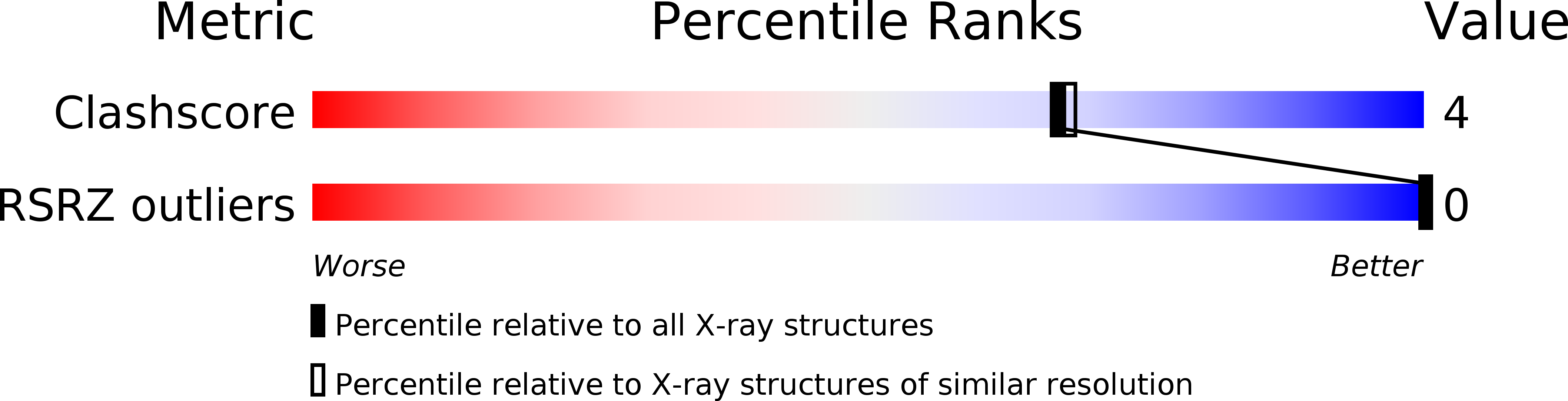Deposition Date
2001-02-20
Release Date
2003-02-11
Last Version Date
2023-08-09
Entry Detail
PDB ID:
1I47
Keywords:
Title:
MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGAATT(MO4)CGCG): THE WATSON-CRICK TYPE AND WOBBLE N4-METHOXYCYTIDINE/GUANOSINE BASE PAIRS IN B-DNA
Method Details:
Experimental Method:
Resolution:
2.10 Å
R-Value Free:
0.25
R-Value Work:
0.21
Space Group:
P 21 21 21