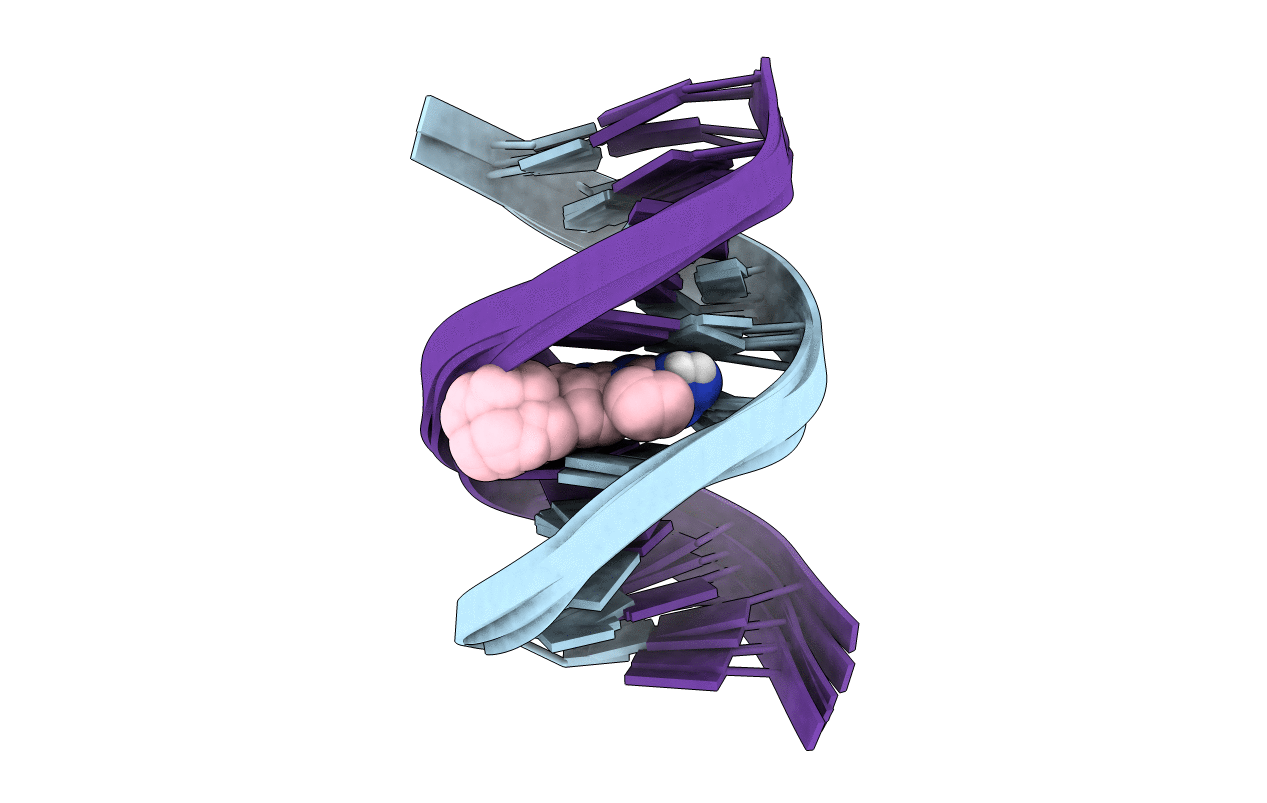
Deposition Date
2001-01-23
Release Date
2001-08-08
Last Version Date
2024-05-22
Entry Detail
PDB ID:
1HZ0
Keywords:
Title:
NMR STRUCTURE OF THE 2-AMINO-1-METHYL-6-PHENYLIMIDAZO[4,5-B]PYRIDINE (PHIP) C8-DEOXYGUANOSINE ADDUCT IN DUPLEX DNA
Biological Source:
Source Organism:
Method Details:
Experimental Method:
Conformers Calculated:
48
Conformers Submitted:
6
Selection Criteria:
structures with the lowest energy


