Deposition Date
2001-01-08
Release Date
2001-01-16
Last Version Date
2024-05-01
Entry Detail
PDB ID:
1HJ6
Keywords:
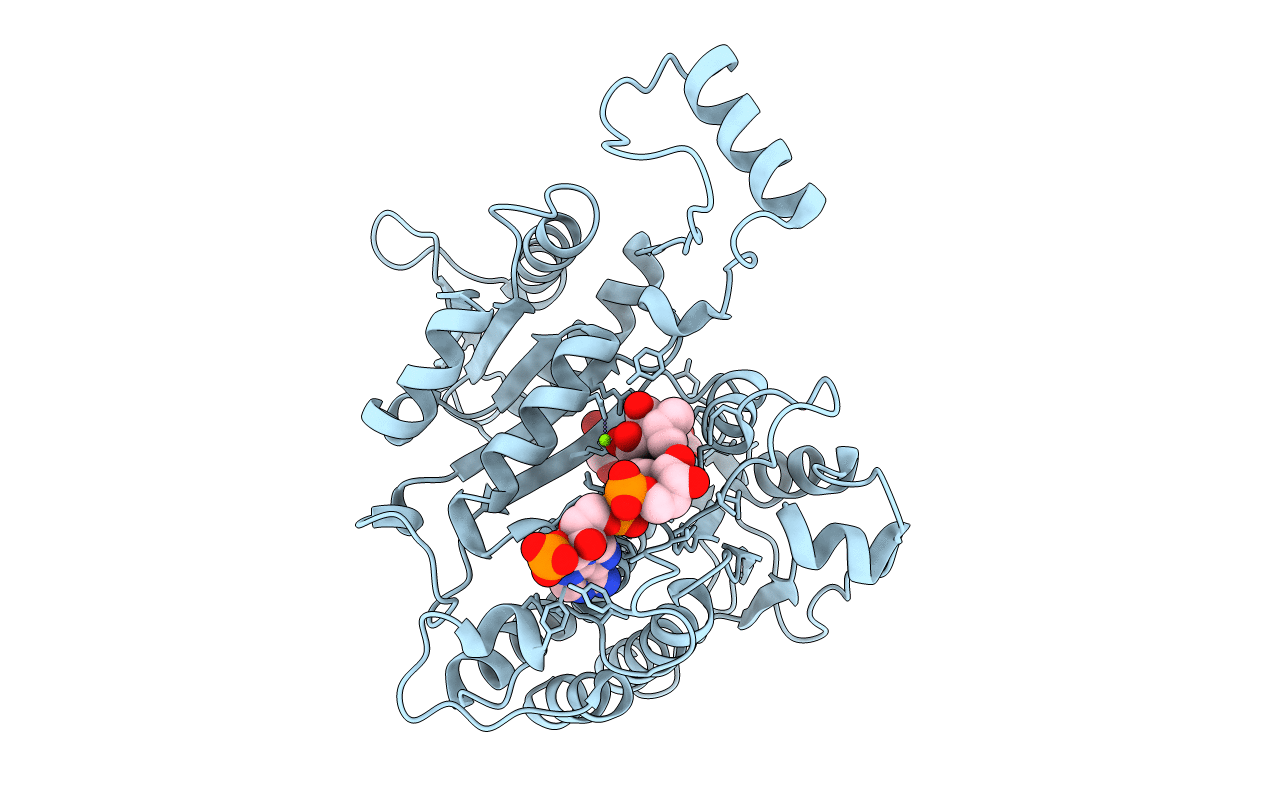
Title:
ISOCITRATE DEHYDROGENASE S113E MUTANT COMPLEXED WITH ISOPROPYLMALATE, NADP+ AND MAGNESIUM (FLASH-COOLED)
Biological Source:
Source Organism:
ESCHERICHIA COLI (Taxon ID: 562)
Host Organism:
Method Details:
Experimental Method:
Resolution:
2.00 Å
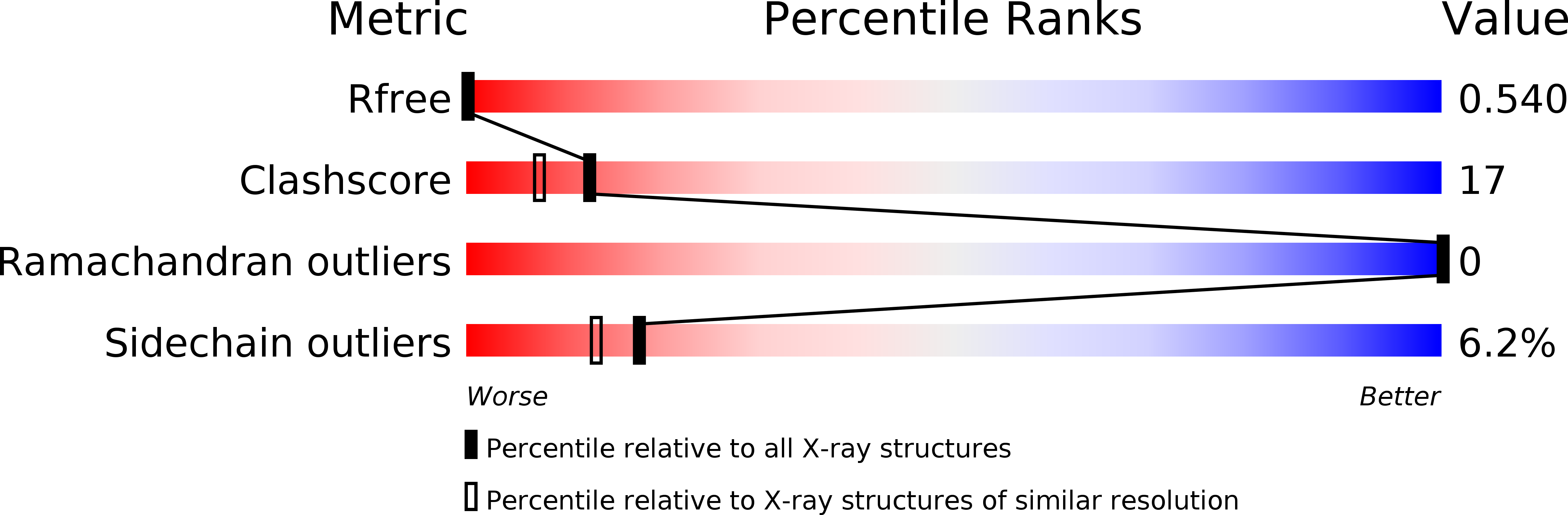
R-Value Free:
0.24
R-Value Work:
0.20
R-Value Observed:
0.20
Space Group:
P 43 21 2