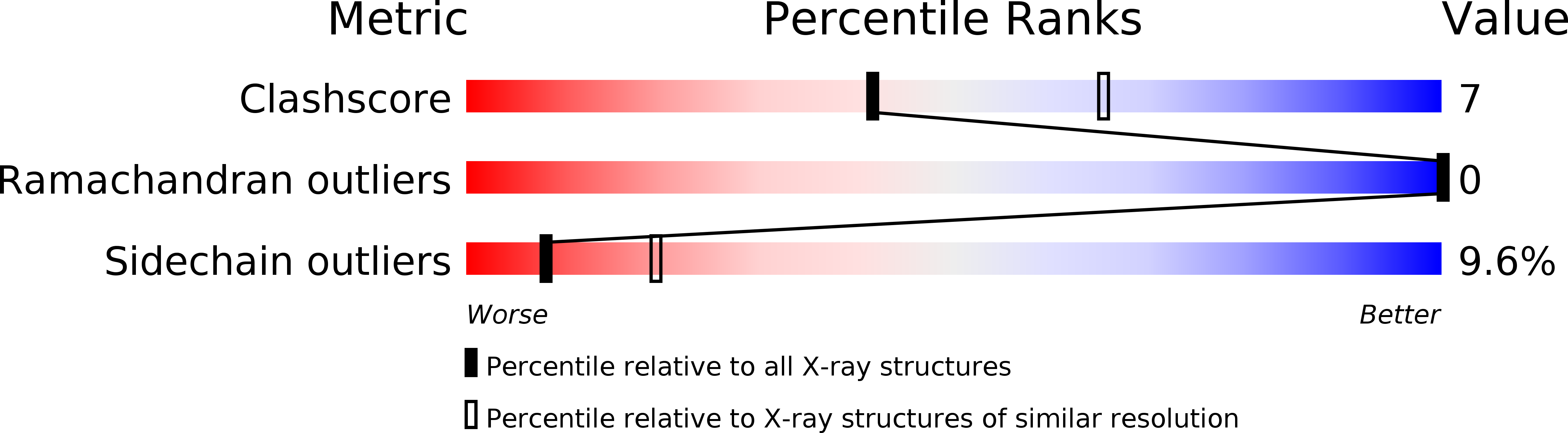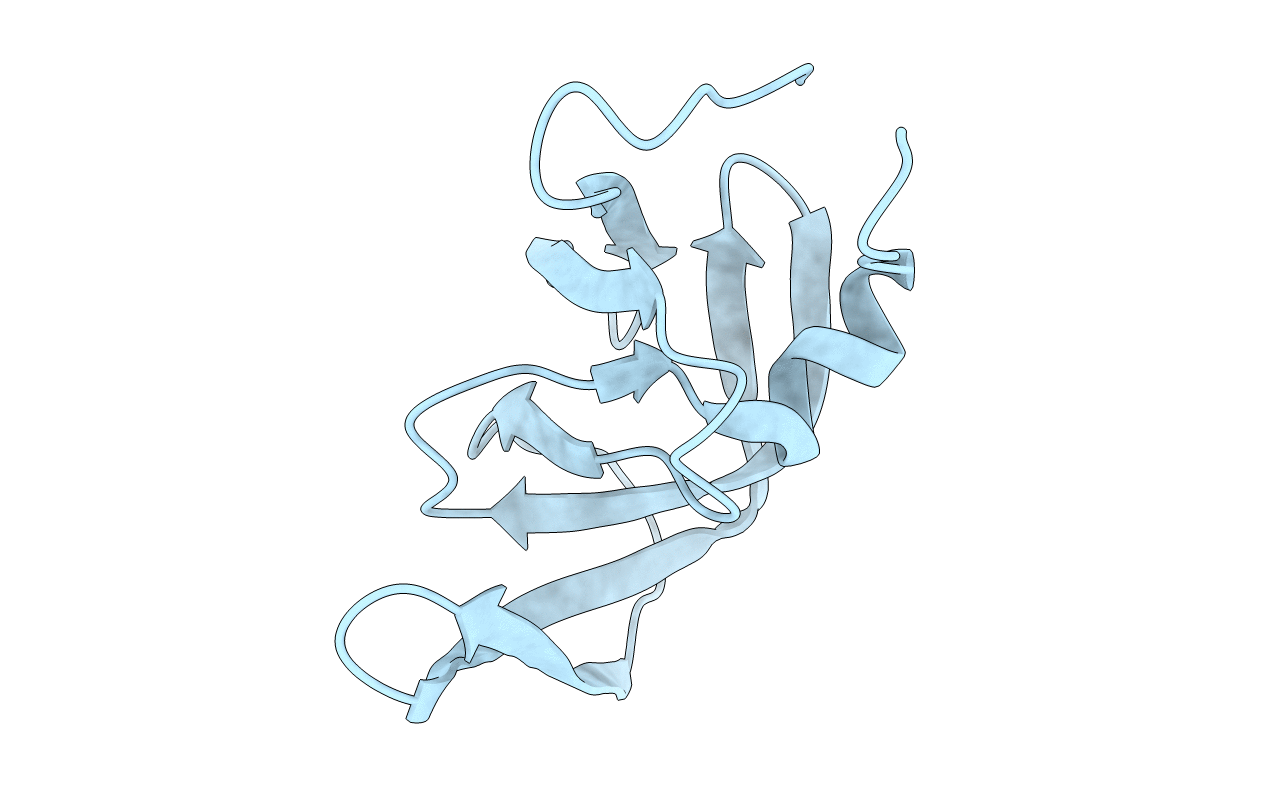
Deposition Date
1992-05-27
Release Date
1992-10-15
Last Version Date
2024-02-07
Entry Detail
PDB ID:
1HHP
Keywords:
Title:
THE THREE-DIMENSIONAL STRUCTURE OF THE ASPARTYL PROTEASE FROM THE HIV-1 ISOLATE BRU
Biological Source:
Source Organism(s):
Expression System(s):
Method Details: