Deposition Date
2001-07-08
Release Date
2004-02-25
Last Version Date
2024-10-23
Entry Detail
PDB ID:
1H7K
Keywords:
Title:
Formation of a tyrosyl radical intermediate in Proteus mirabilis catalase by directed mutagenesis and consequences for nucleotide reactivity
Biological Source:
Source Organism(s):
PROTEUS MIRABILIS (Taxon ID: 584)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.40 Å
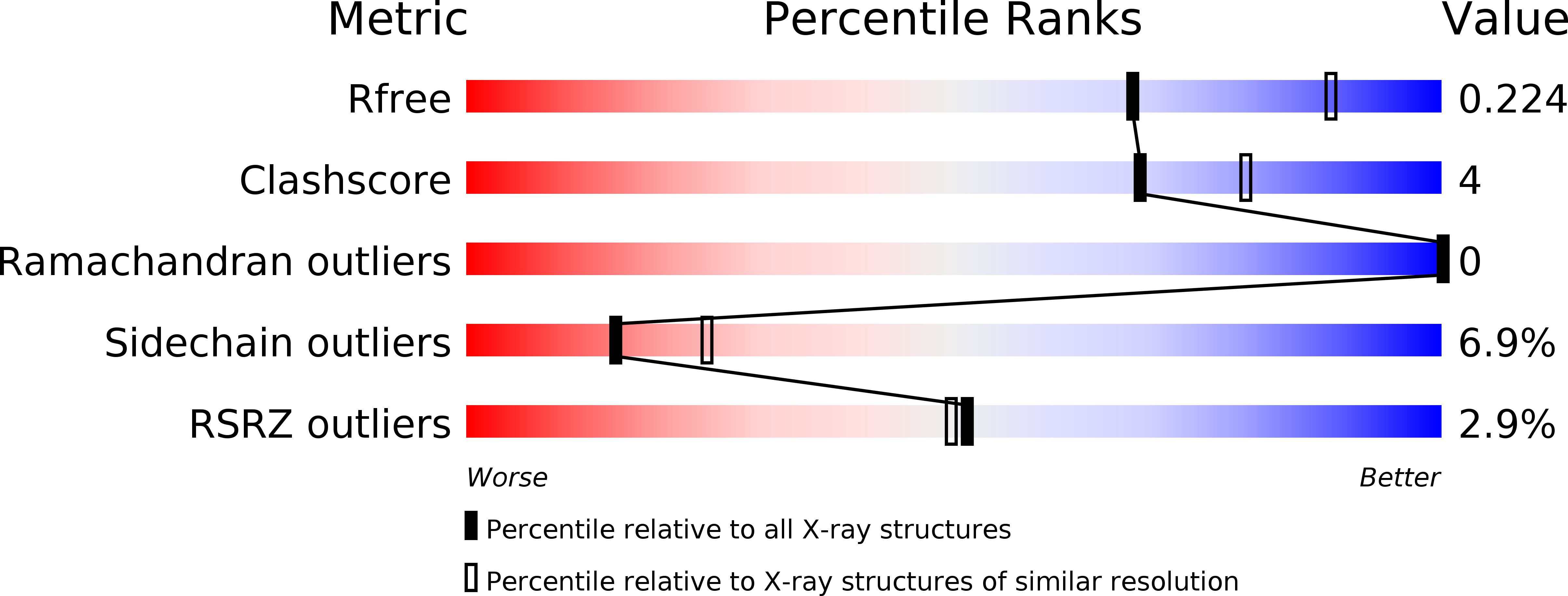
R-Value Free:
0.24
R-Value Work:
0.23
R-Value Observed:
0.23
Space Group:
P 62 2 2