Deposition Date
2001-06-12
Release Date
2001-11-28
Last Version Date
2023-12-13
Entry Detail
PDB ID:
1H6E
Keywords:
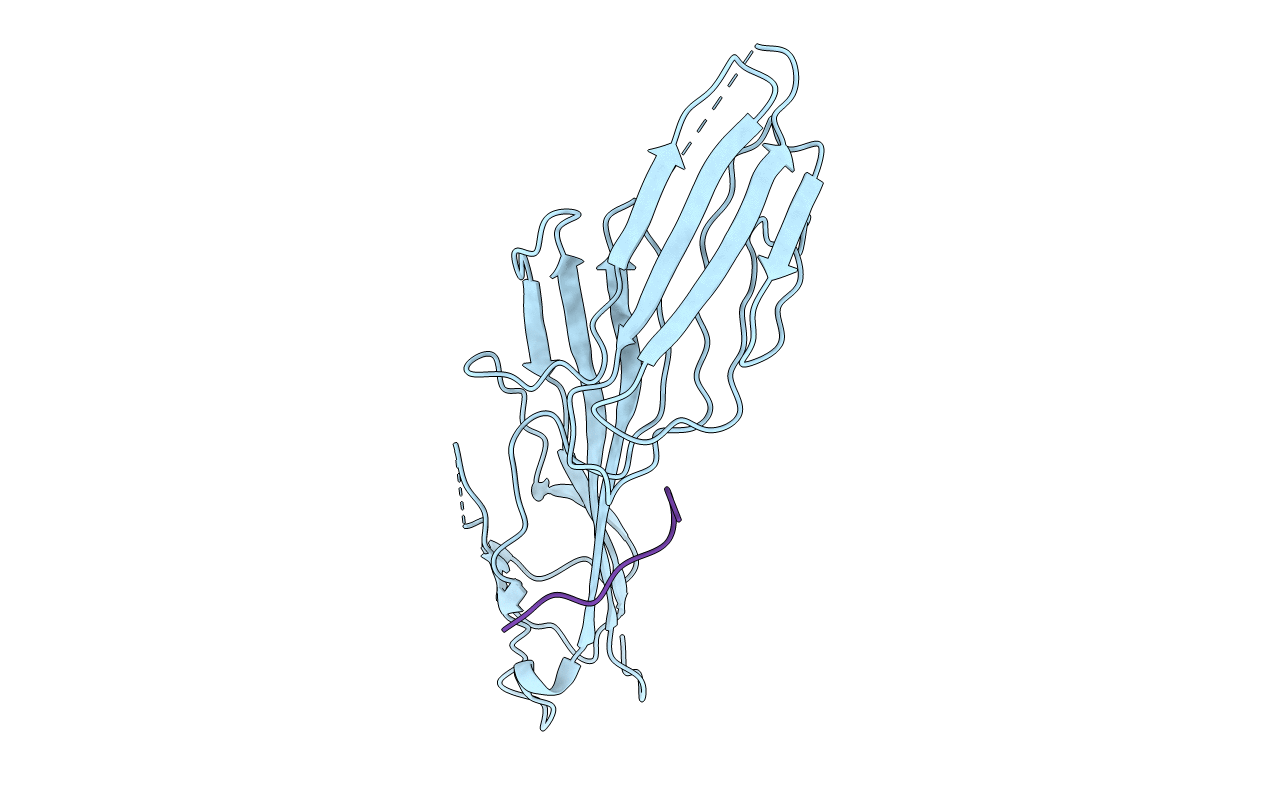
Title:
MU2 ADAPTIN SUBUNIT (AP50) OF AP2 ADAPTOR (SECOND DOMAIN), COMPLEXED WITH CTLA-4 INTERNALIZATION PEPTIDE TTGVYVKMPPT
Biological Source:
Source Organism:
HOMO SAPIENS (Taxon ID: 9606)
Host Organism:
Method Details:
Experimental Method:
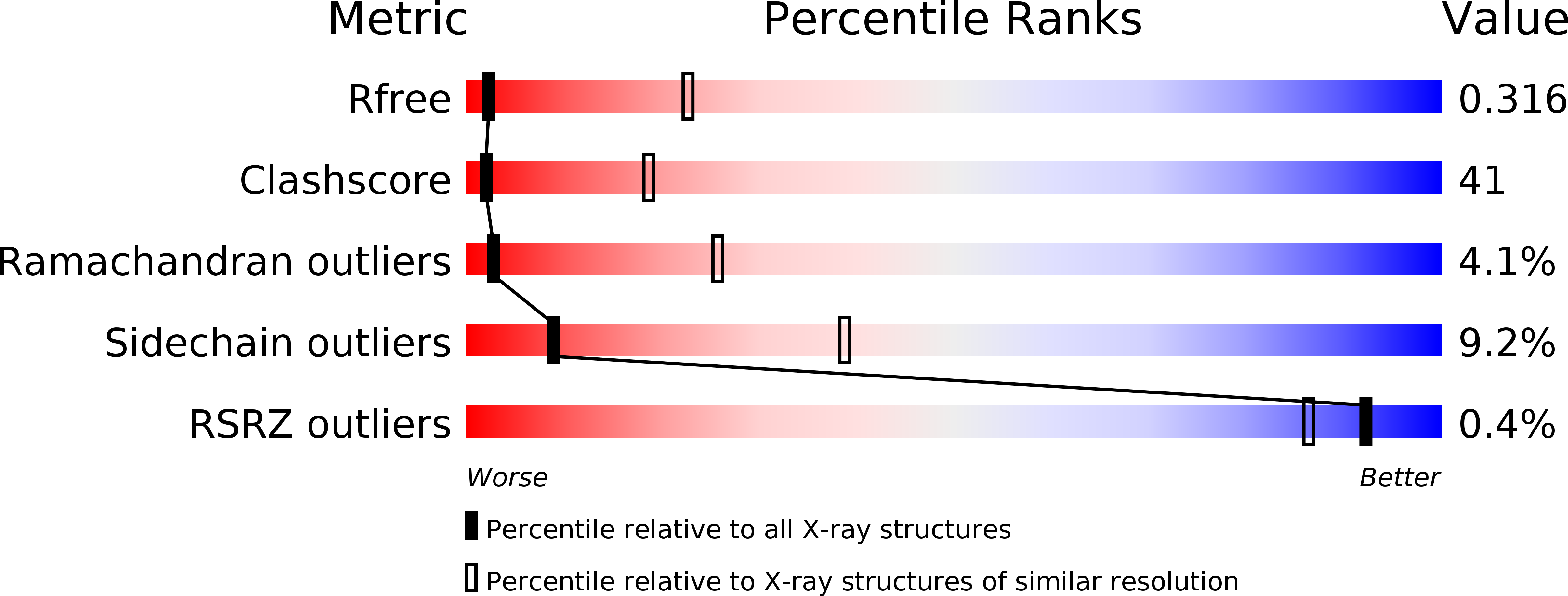
Resolution:
3.60 Å
R-Value Free:
0.31
R-Value Work:
0.27
Space Group:
P 64