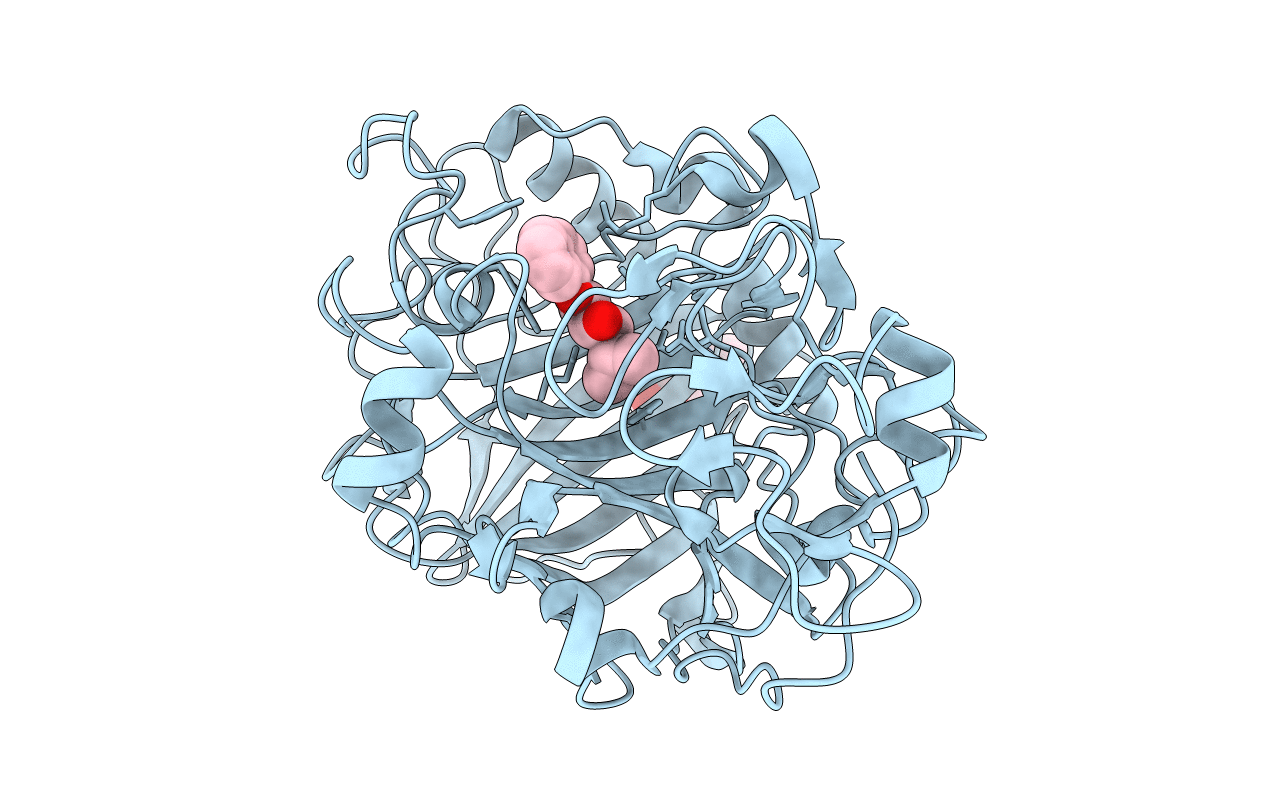
Deposition Date
2002-10-03
Release Date
2003-04-03
Last Version Date
2024-11-13
Entry Detail
PDB ID:
1H46
Keywords:
Title:
The catalytic module of Cel7D from Phanerochaete chrysosporium as a chiral selector: Structural studies of its complex with the b-blocker (R)-propranolol
Biological Source:
Source Organism(s):
PHANEROCHAETE CHRYSOSPORIUM (Taxon ID: 5306)
Method Details:
Experimental Method:
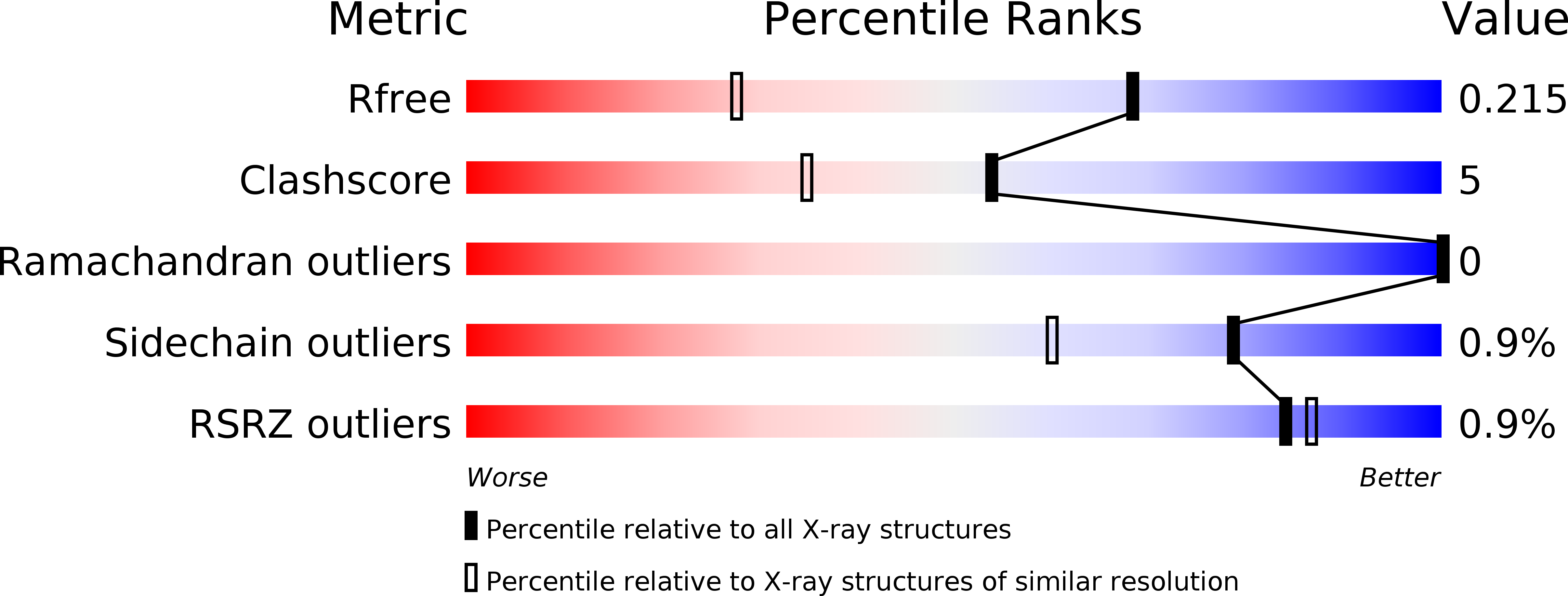
Resolution:
1.52 Å
R-Value Free:
0.21
R-Value Work:
0.16
R-Value Observed:
0.17
Space Group:
C 1 2 1