Deposition Date
2001-11-05
Release Date
2002-01-01
Last Version Date
2024-11-13
Entry Detail
PDB ID:
1GPI
Keywords:
Title:
Cellobiohydrolase Cel7D (CBH 58) from Phanerochaete chrysosporium. Catalytic module at 1.32 Angstrom resolution
Biological Source:
Source Organism(s):
PHANEROCHAETE CHRYSOSPORIUM (Taxon ID: 5306)
Method Details:
Experimental Method:
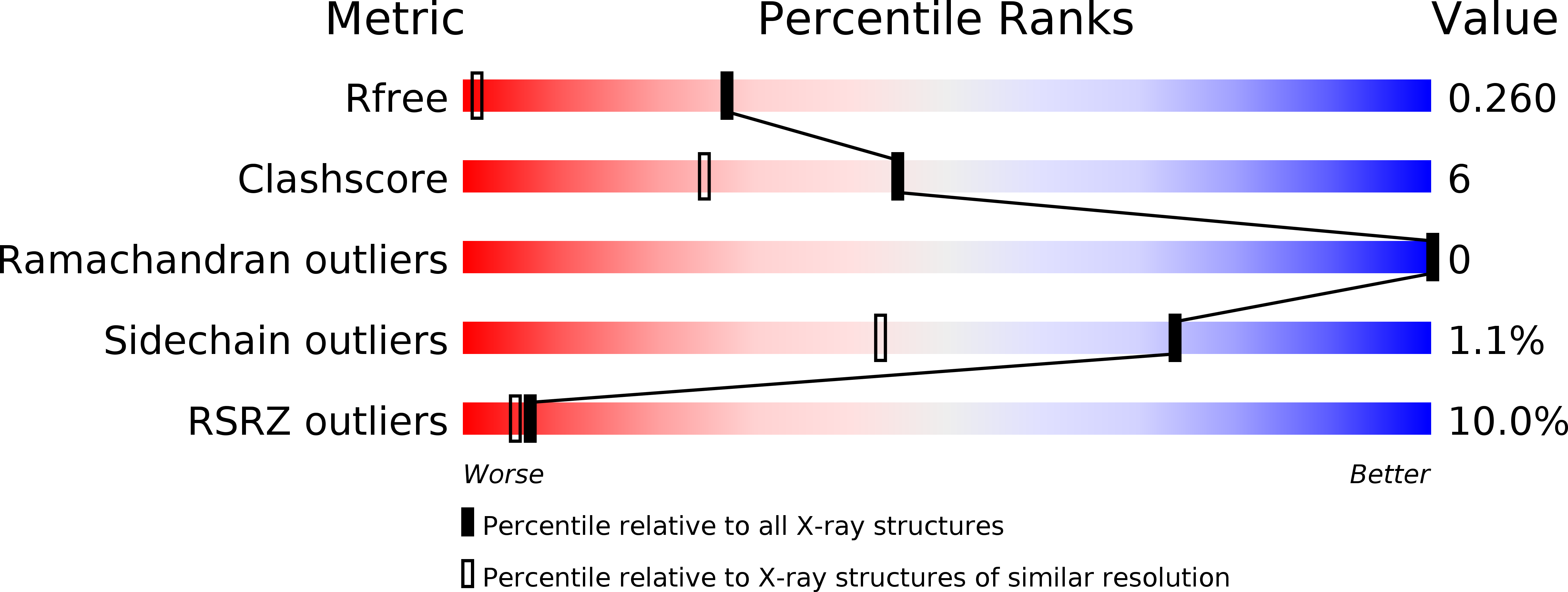
Resolution:
1.32 Å
R-Value Free:
0.24
R-Value Work:
0.21
R-Value Observed:
0.21
Space Group:
C 1 2 1