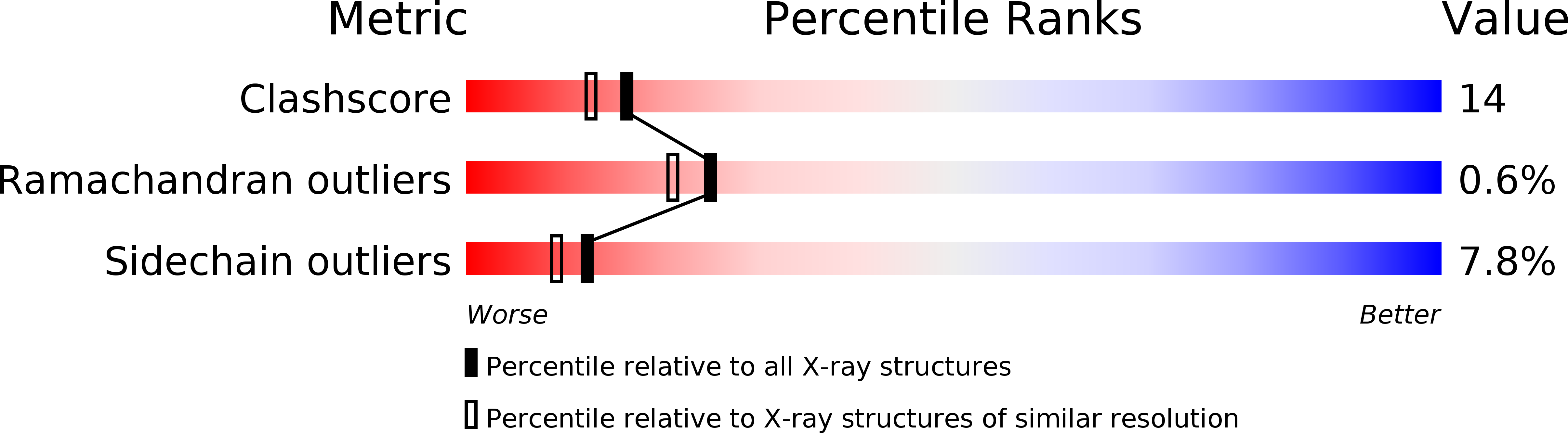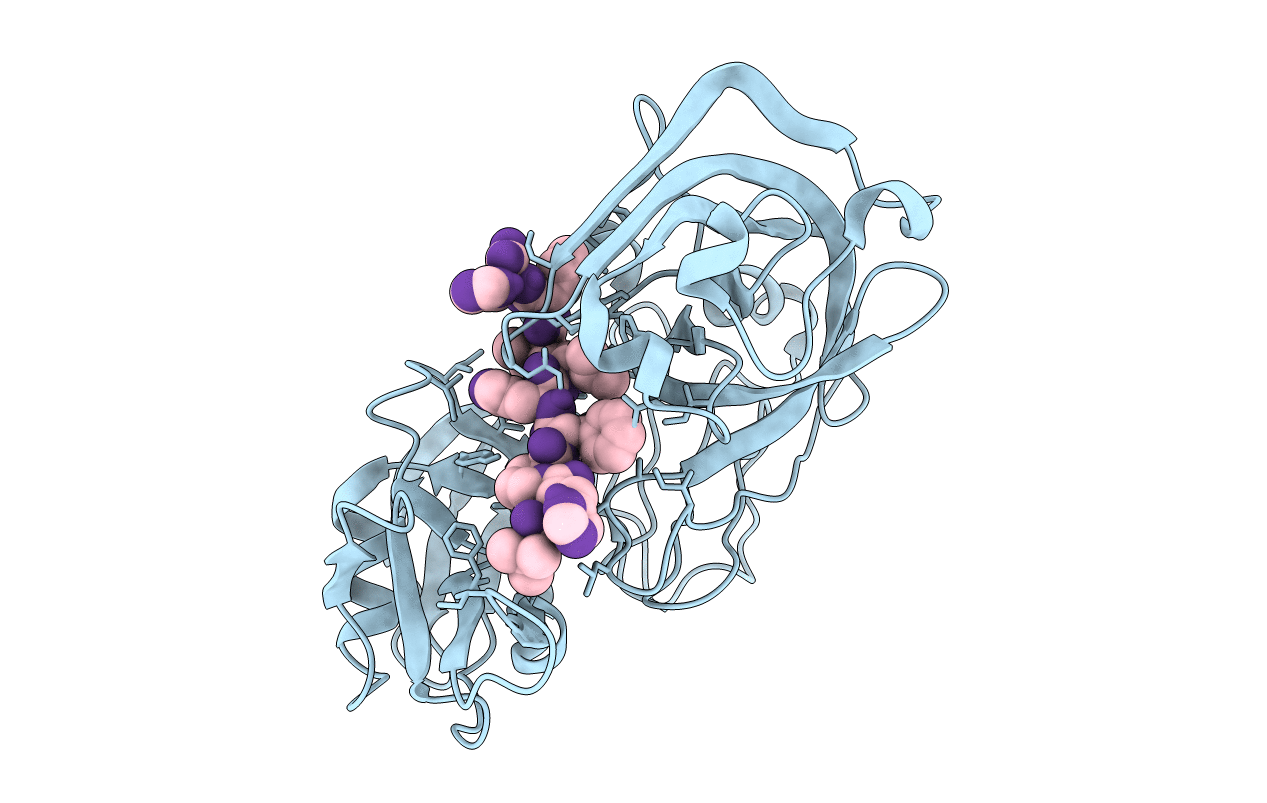
Deposition Date
2001-08-20
Release Date
2001-11-20
Last Version Date
2023-11-15
Entry Detail
PDB ID:
1GKT
Keywords:
Title:
Neutron Laue diffraction structure of endothiapepsin complexed with transition state analogue inhibitor H261
Biological Source:
Source Organism(s):
synthetic construct (Taxon ID: 32630)
CRYPHONECTRIA PARASITICA (Taxon ID: 5116)
CRYPHONECTRIA PARASITICA (Taxon ID: 5116)
Method Details:
Experimental Method:
Resolution:
2.10 Å
R-Value Free:
0.27
R-Value Observed:
0.23
Space Group:
P 1 21 1