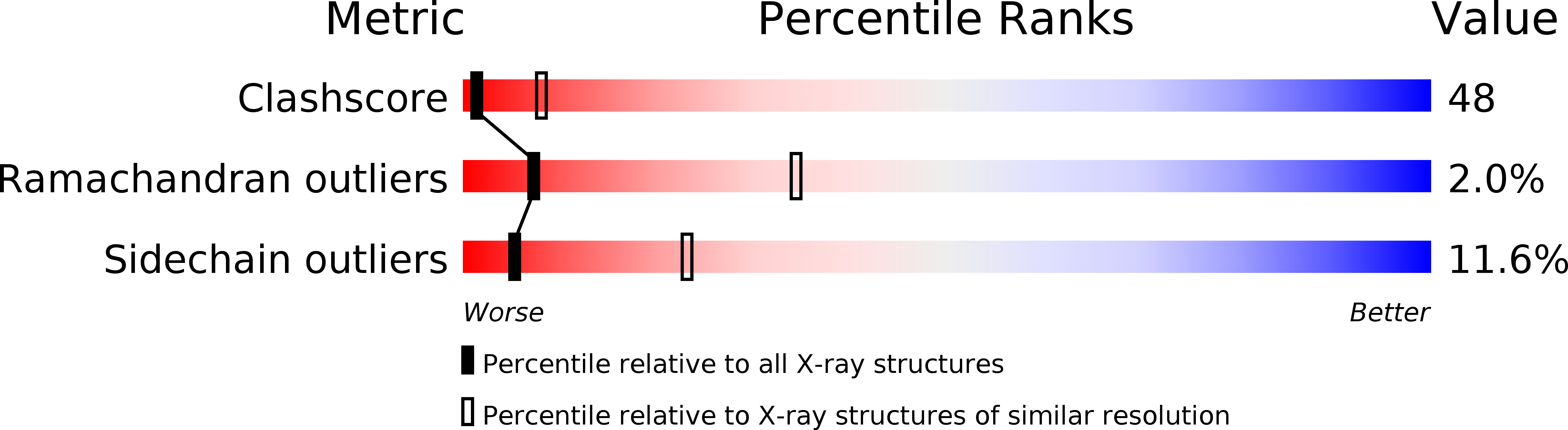Deposition Date
2000-10-16
Release Date
2001-01-31
Last Version Date
2023-08-09
Entry Detail
PDB ID:
1G21
Keywords:
Title:
MGATP-BOUND AND NUCLEOTIDE-FREE STRUCTURES OF A NITROGENASE PROTEIN COMPLEX BETWEEN LEU127DEL-FE PROTEIN AND THE MOFE PROTEIN
Biological Source:
Source Organism(s):
Azotobacter vinelandii (Taxon ID: 354)
Method Details:
Experimental Method:
Resolution:
3.00 Å
R-Value Free:
0.28
R-Value Work:
0.23
R-Value Observed:
0.24
Space Group:
P 21 21 21