Deposition Date
2000-10-03
Release Date
2000-11-08
Last Version Date
2024-10-30
Entry Detail
PDB ID:
1FYT
Keywords:
Title:
CRYSTAL STRUCTURE OF A COMPLEX OF A HUMAN ALPHA/BETA-T CELL RECEPTOR, INFLUENZA HA ANTIGEN PEPTIDE, AND MHC CLASS II MOLECULE, HLA-DR1
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
H3N2 subtype (Taxon ID: 119210)
H3N2 subtype (Taxon ID: 119210)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.60 Å
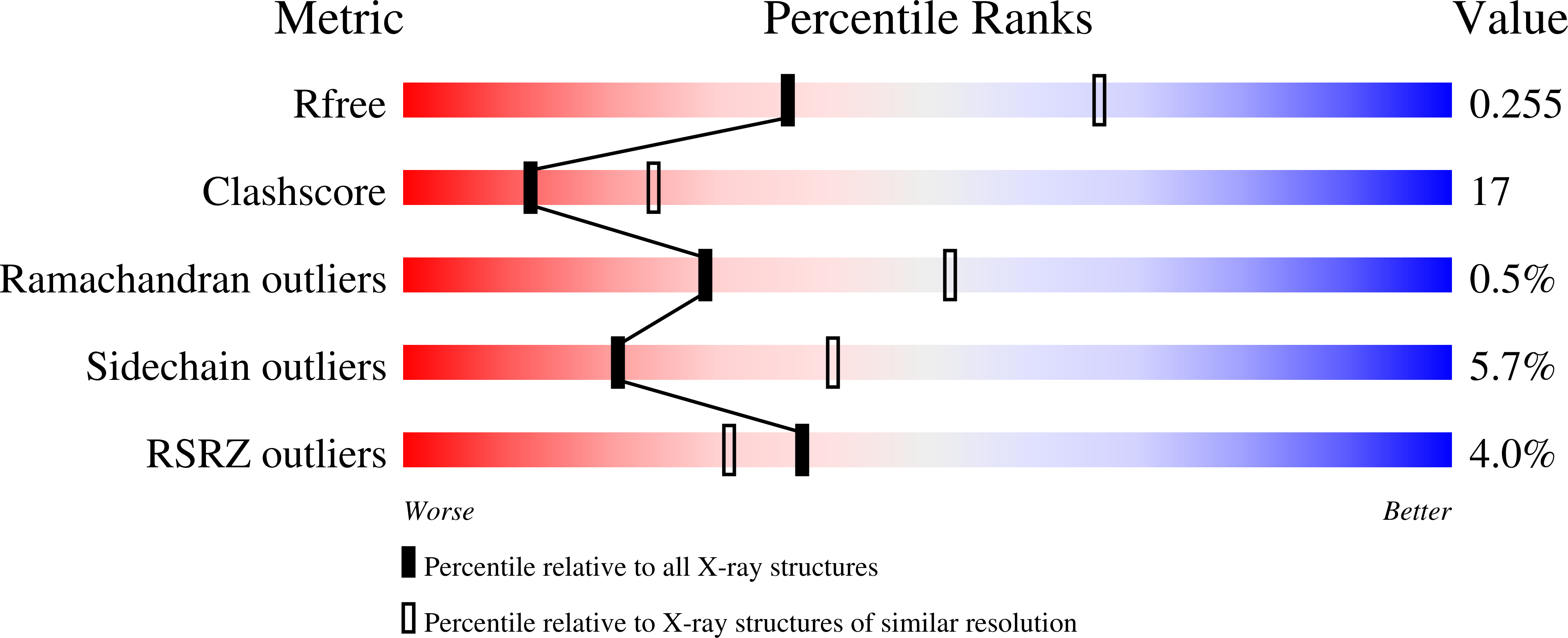
R-Value Free:
0.25
R-Value Work:
0.22
Space Group:
C 1 2 1