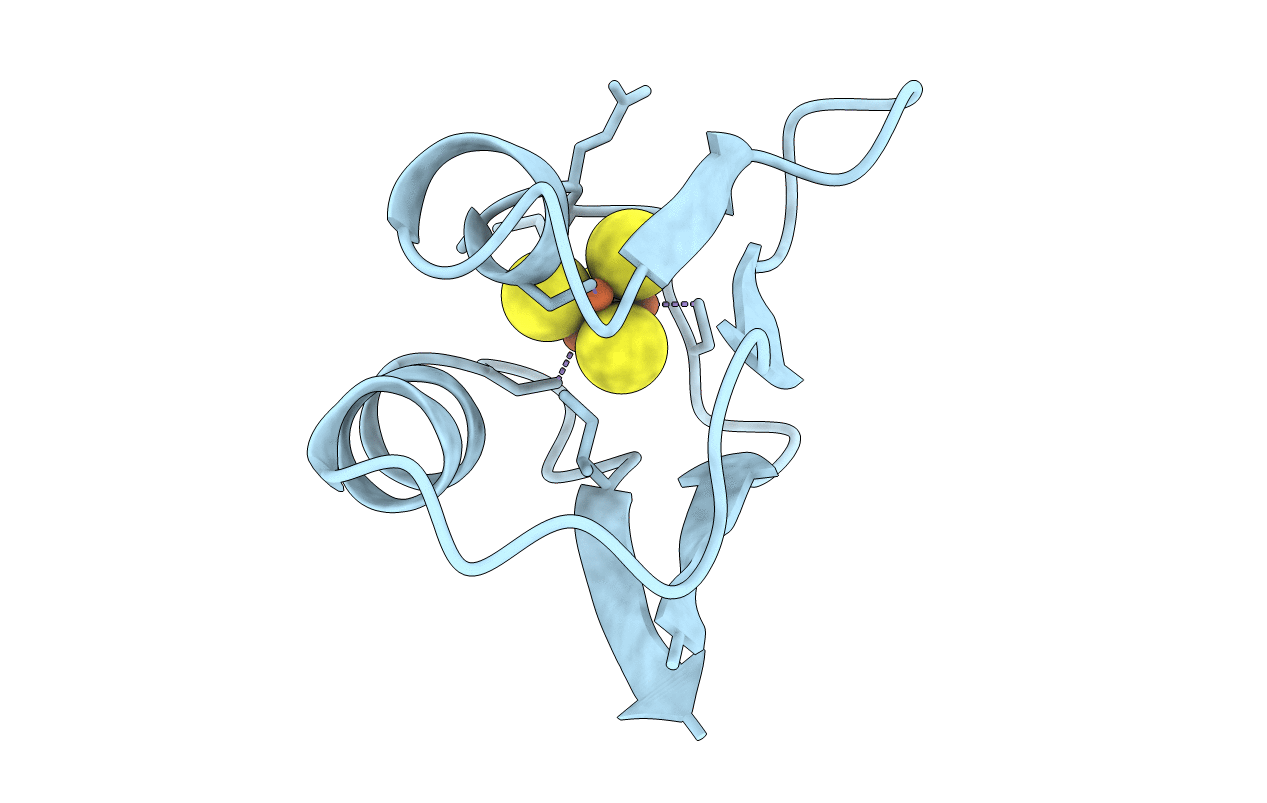
Deposition Date
1991-04-08
Release Date
1993-04-15
Last Version Date
2025-03-26
Entry Detail
PDB ID:
1FXD
Keywords:
Title:
REFINED CRYSTAL STRUCTURE OF FERREDOXIN II FROM DESULFOVIBRIO GIGAS AT 1.7 ANGSTROMS
Biological Source:
Source Organism(s):
Desulfovibrio gigas (Taxon ID: 879)
Method Details:
Experimental Method:
Resolution:
1.70 Å
R-Value Observed:
0.15
Space Group:
C 1 2 1


