Deposition Date
1993-04-14
Release Date
1994-05-31
Last Version Date
2024-02-07
Entry Detail
PDB ID:
1FRD
Keywords:
Title:
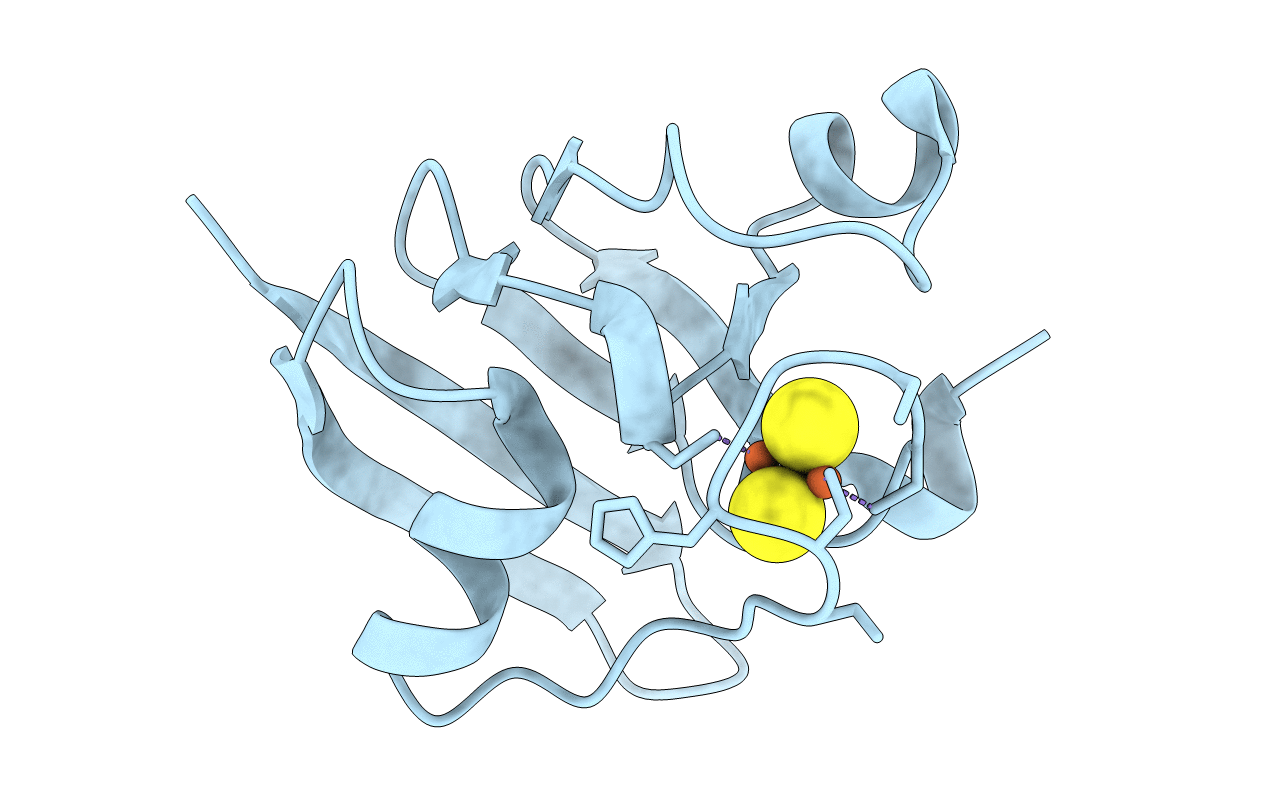
MOLECULAR STRUCTURE OF THE OXIDIZED, RECOMBINANT, HETEROCYST (2FE-2S) FERREDOXIN FROM ANABAENA 7120 DETERMINED TO 1.7 ANGSTROMS RESOLUTION
Biological Source:
Source Organism(s):
Nostoc sp. (Taxon ID: 103690)
Method Details: