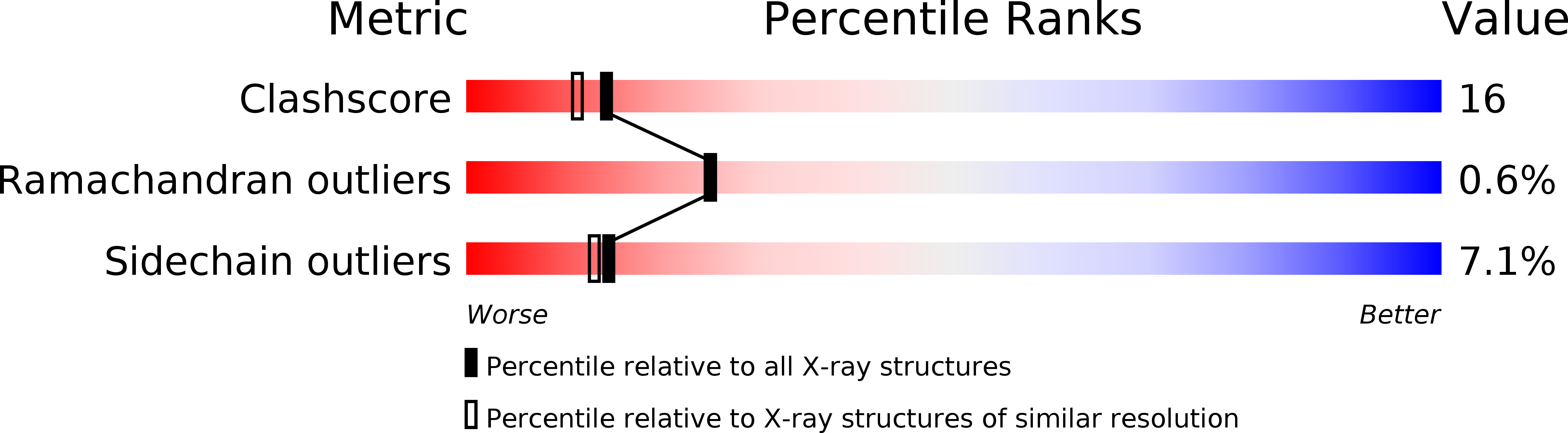Deposition Date
2000-08-08
Release Date
2000-12-15
Last Version Date
2024-02-07
Entry Detail
PDB ID:
1FJX
Keywords:
Title:
STRUCTURE OF TERNARY COMPLEX OF HHAI METHYLTRANSFERASE MUTANT (T250G) IN COMPLEX WITH DNA AND ADOHCY
Biological Source:
Source Organism(s):
Haemophilus haemolyticus (Taxon ID: 726)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.26 Å
R-Value Free:
0.28
R-Value Work:
0.20
Space Group:
H 3 2