Deposition Date
2000-05-29
Release Date
2000-06-08
Last Version Date
2024-05-22
Entry Detail
PDB ID:
1F2R
Keywords:
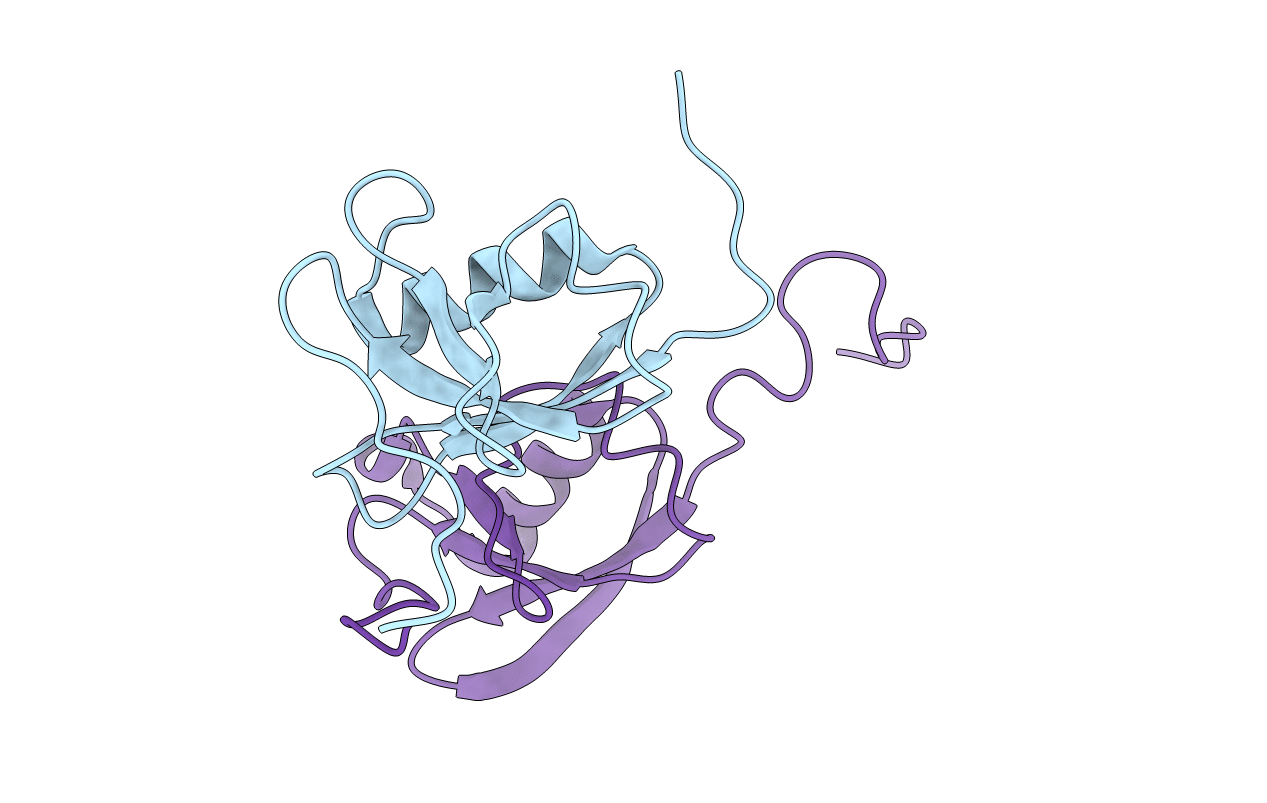
Title:
NMR STRUCTURE OF THE HETERODIMERIC COMPLEX BETWEEN CAD DOMAINS OF CAD AND ICAD
Biological Source:
Source Organism(s):
Mus musculus (Taxon ID: 10090)
Expression System(s):
Method Details:
Experimental Method:
Conformers Submitted:
1


