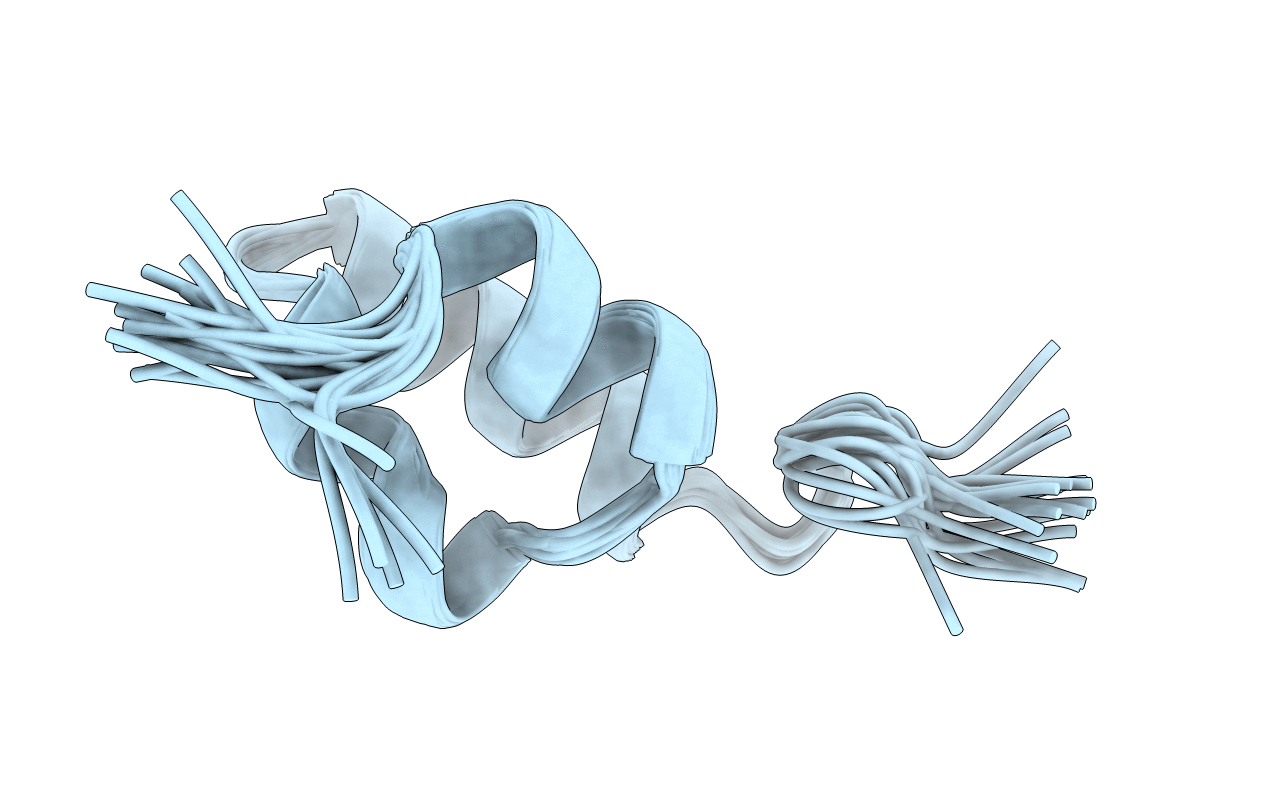
Deposition Date
1994-02-14
Release Date
1994-10-15
Last Version Date
2024-10-09
Entry Detail
PDB ID:
1ERD
Keywords:
Title:
THE NMR SOLUTION STRUCTURE OF THE PHEROMONE ER-2 FROM THE CILIATED PROTOZOAN EUPLOTES RAIKOVI
Biological Source:
Source Organism(s):
Euplotes raikovi (Taxon ID: 5938)
Method Details:
Experimental Method:
Conformers Submitted:
20