Deposition Date
2000-03-09
Release Date
2000-09-01
Last Version Date
2024-02-07
Entry Detail
PDB ID:
1EKU
Keywords:
Title:
CRYSTAL STRUCTURE OF A BIOLOGICALLY ACTIVE SINGLE CHAIN MUTANT OF HUMAN IFN-GAMMA
Biological Source:
Source Organism:
Homo sapiens (Taxon ID: 9606)
Host Organism:
Method Details:
Experimental Method:
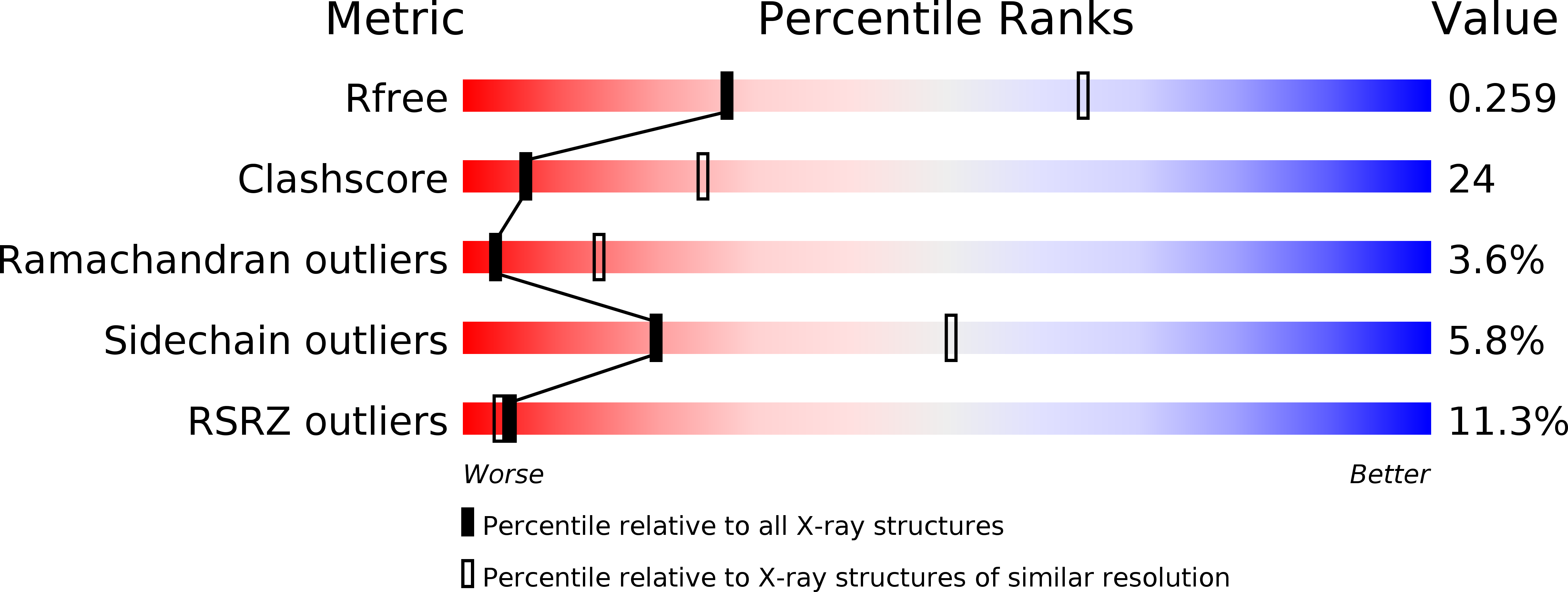
Resolution:
2.90 Å
R-Value Free:
0.26
R-Value Work:
0.24
R-Value Observed:
0.25
Space Group:
H 3 2