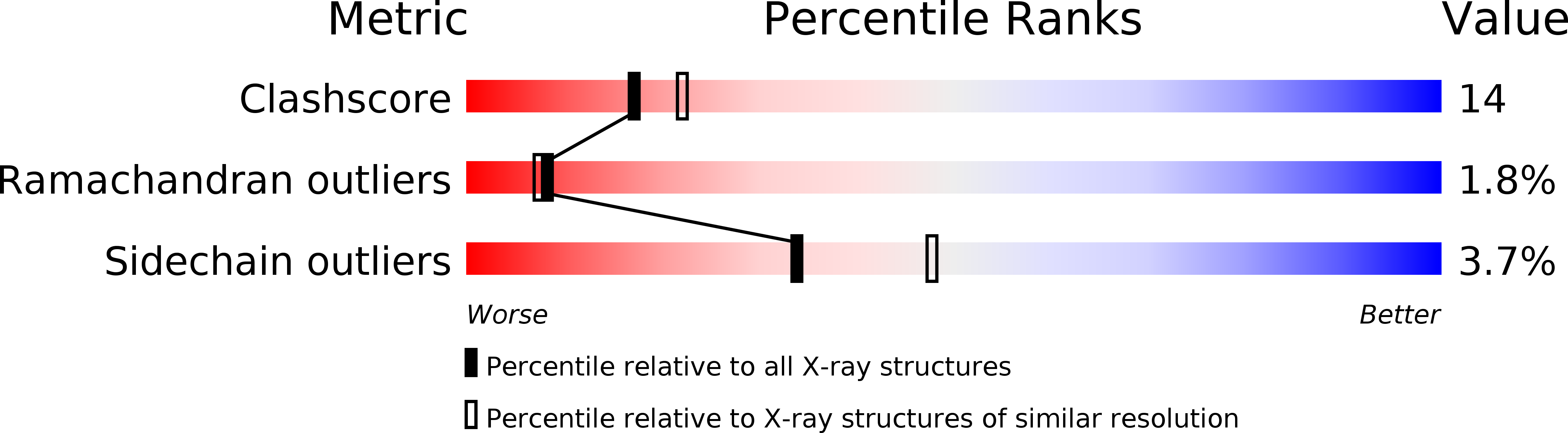Deposition Date
2000-03-03
Release Date
2000-03-22
Last Version Date
2024-11-13
Entry Detail
PDB ID:
1EJO
Keywords:
Title:
FAB FRAGMENT OF NEUTRALISING MONOCLONAL ANTIBODY 4C4 COMPLEXED WITH G-H LOOP FROM FMDV.
Biological Source:
Source Organism(s):
Mus musculus (Taxon ID: 10090)
Method Details:
Experimental Method:
Resolution:
2.30 Å
R-Value Free:
0.26
R-Value Work:
0.24
Space Group:
P 21 21 21