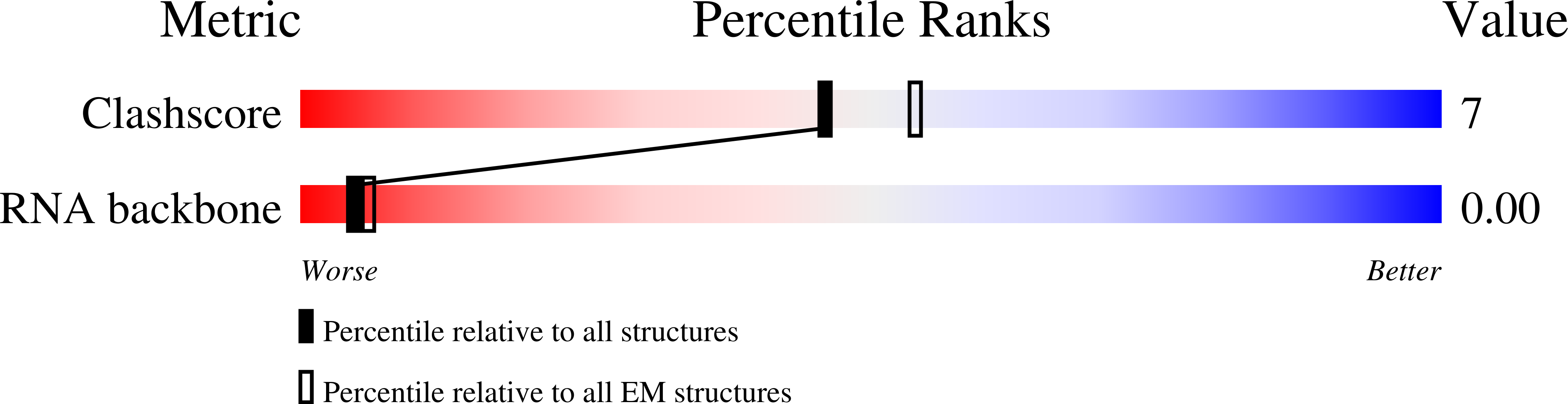
Deposition Date
2000-02-11
Release Date
2000-03-06
Last Version Date
2024-02-07
Entry Detail
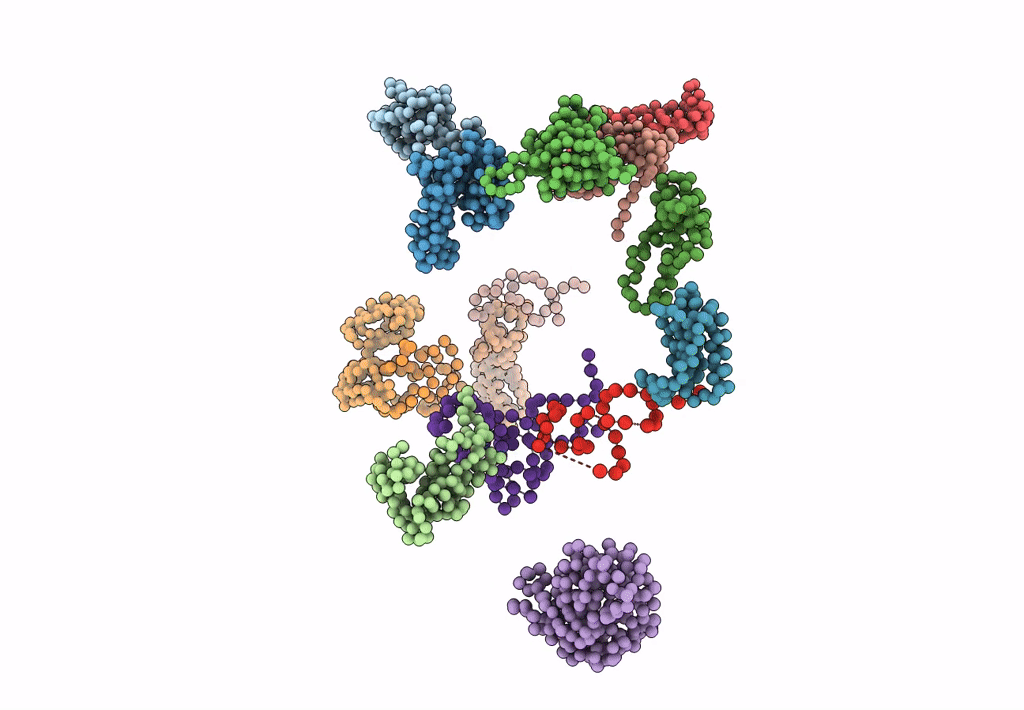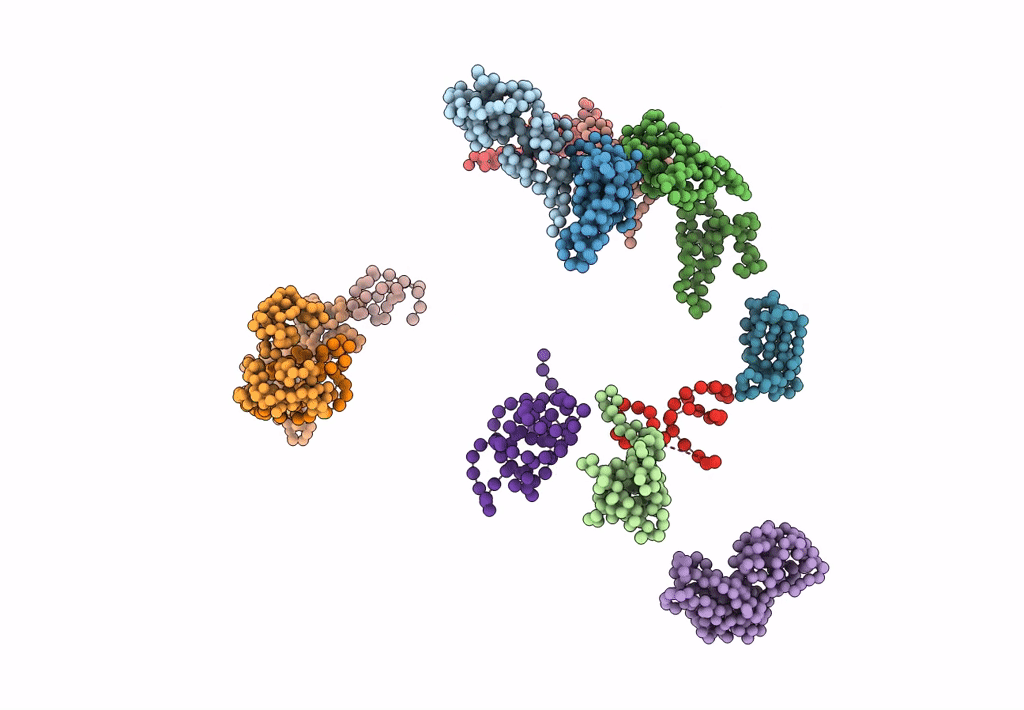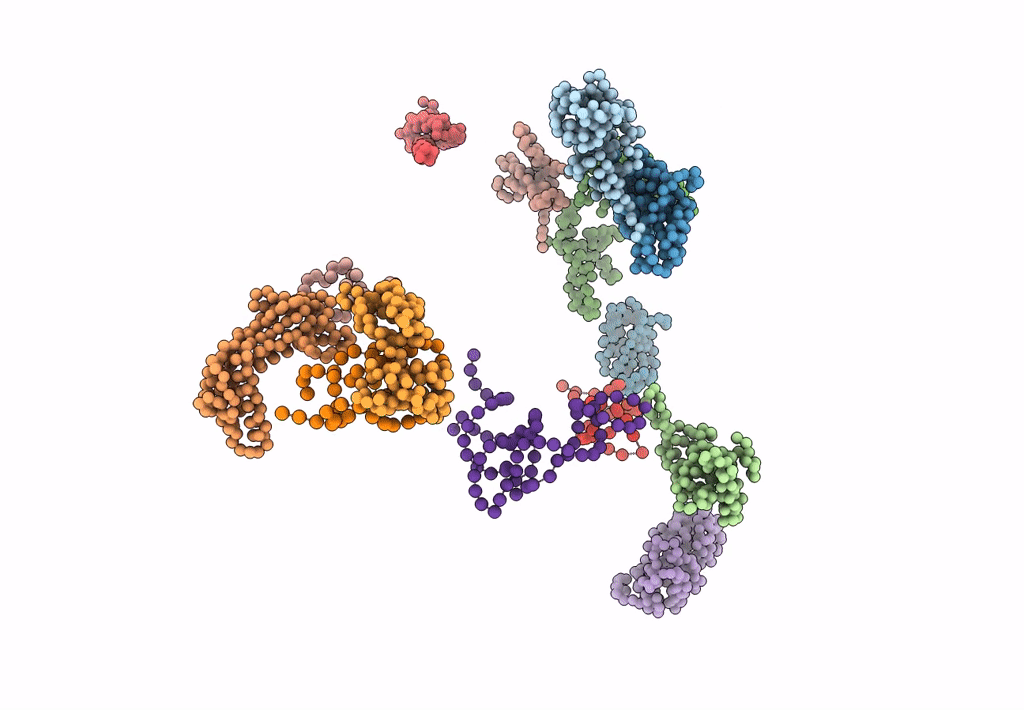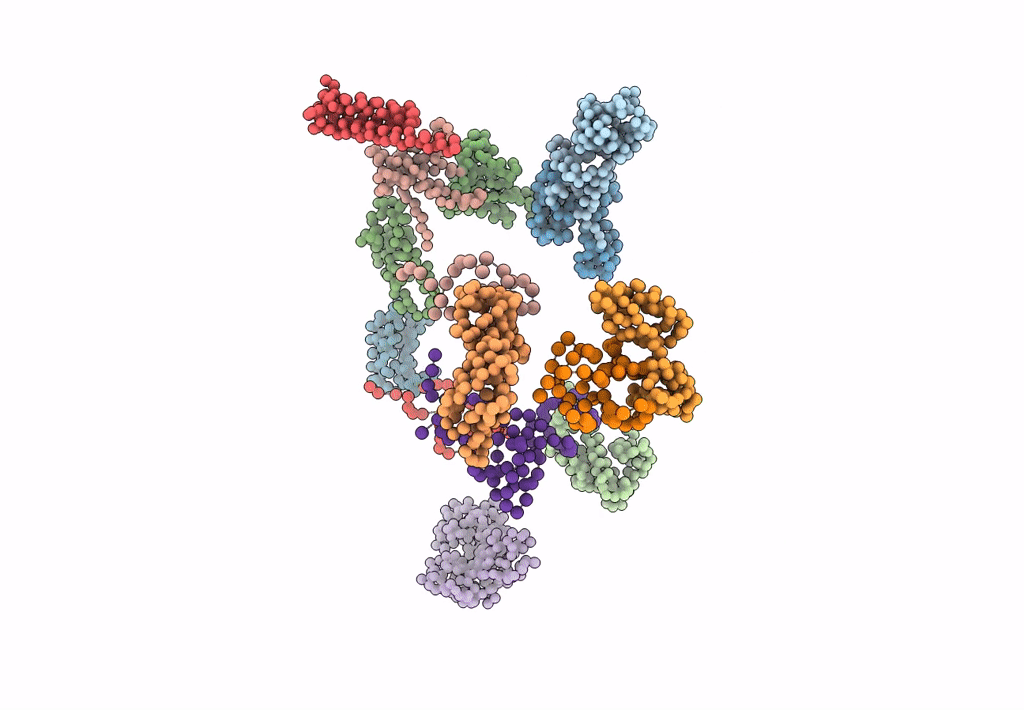
PDB ID:
1EG0
Keywords:
Title:
FITTING OF COMPONENTS WITH KNOWN STRUCTURE INTO AN 11.5 A CRYO-EM MAP OF THE E.COLI 70S RIBOSOME
Biological Source:
Source Organism(s):
Escherichia coli (Taxon ID: 562)
Method Details:
Experimental Method:
Resolution:
11.50 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE