Deposition Date
1987-05-29
Release Date
1987-07-16
Last Version Date
2024-02-07
Entry Detail
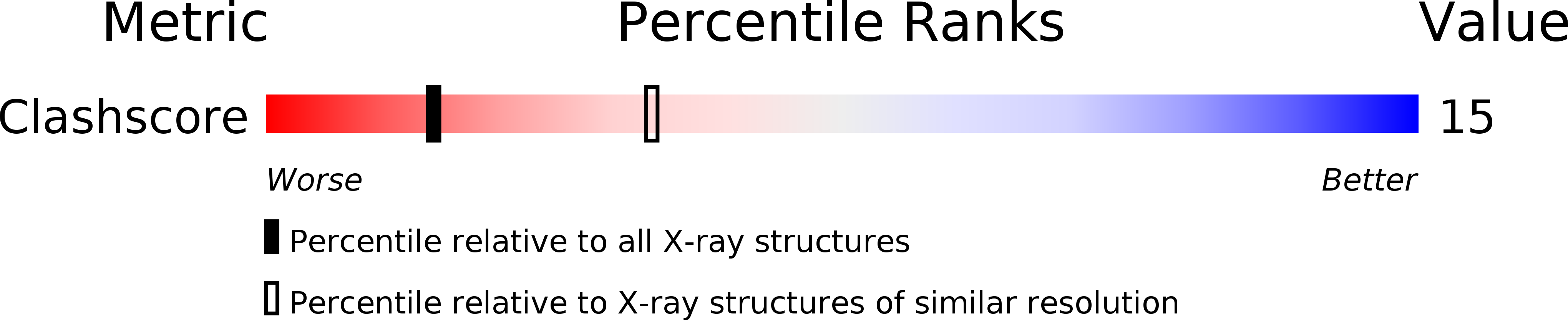
PDB ID:
1EFM
Keywords:
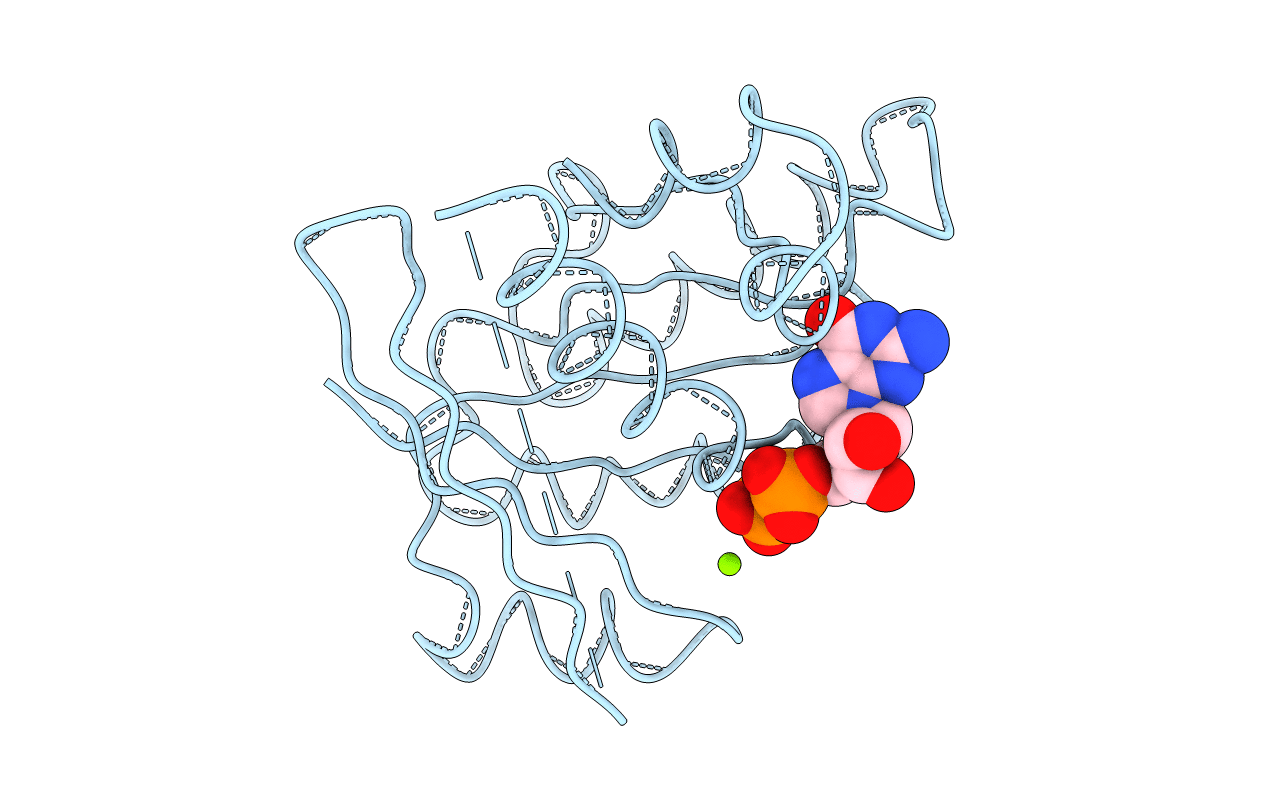
Title:
STRUCTURE OF THE GDP DOMAIN OF EF-TU AND LOCATION OF THE AMINO ACIDS HOMOLOGOUS TO RAS ONCOGENE PROTEINS
Biological Source:
Source Organism(s):
Escherichia coli (Taxon ID: 562)
Method Details: