Deposition Date
1993-05-13
Release Date
1993-10-31
Last Version Date
2024-02-07
Entry Detail
PDB ID:
1EDE
Keywords:
Title:
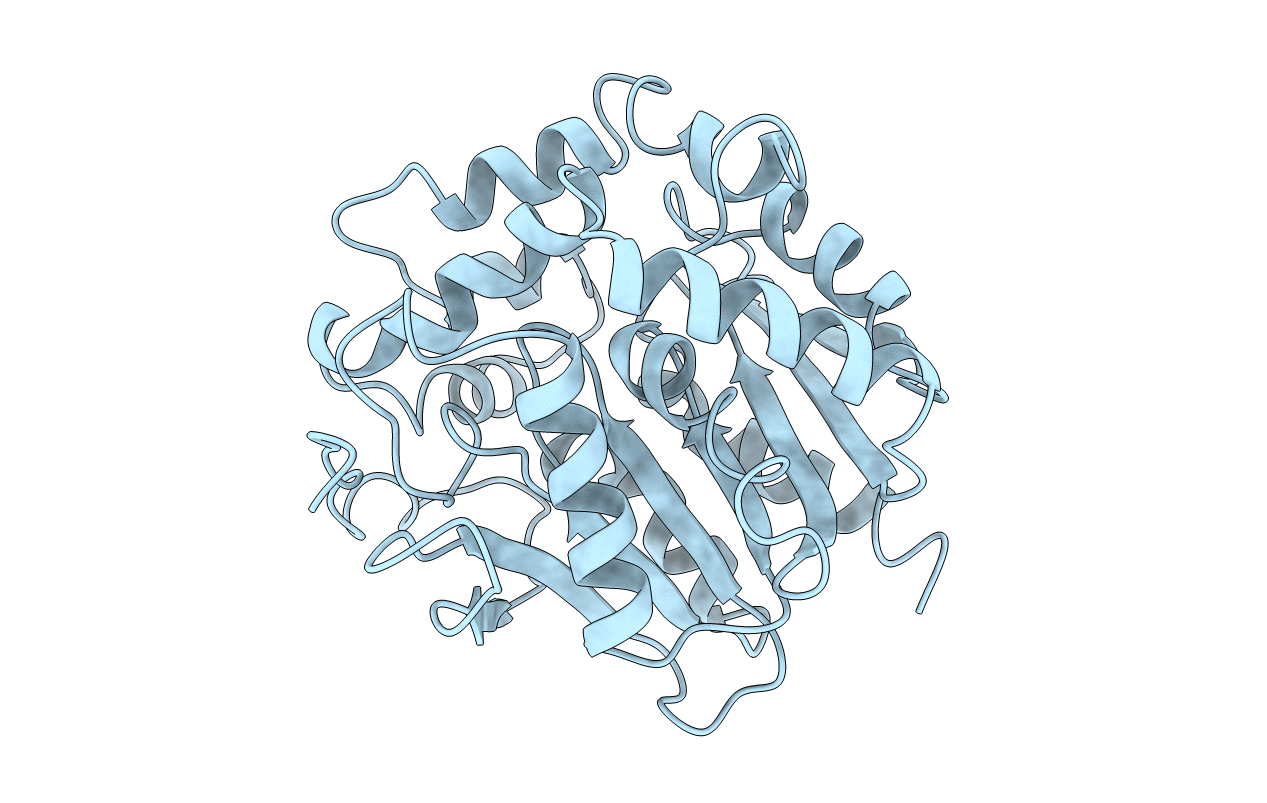
REFINED X-RAY STRUCTURES OF HALOALKANE DEHALOGENASE AT PH 6.2 AND PH 8.2 AND IMPLICATIONS FOR THE REACTION MECHANISM
Biological Source:
Source Organism(s):
Xanthobacter autotrophicus (Taxon ID: 280)
Method Details:
Experimental Method:
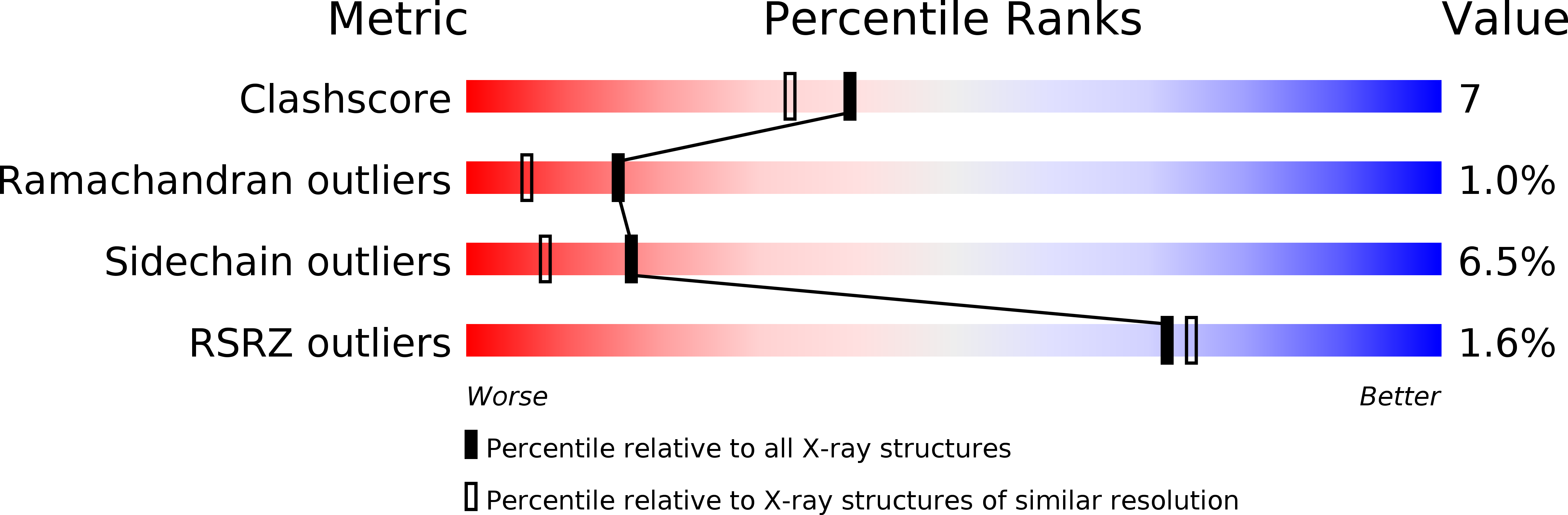
Resolution:
1.90 Å
R-Value Observed:
0.16
Space Group:
P 21 21 2