Deposition Date
1995-05-05
Release Date
1995-07-31
Last Version Date
2024-02-07
Entry Detail
PDB ID:
1ECL
Keywords:
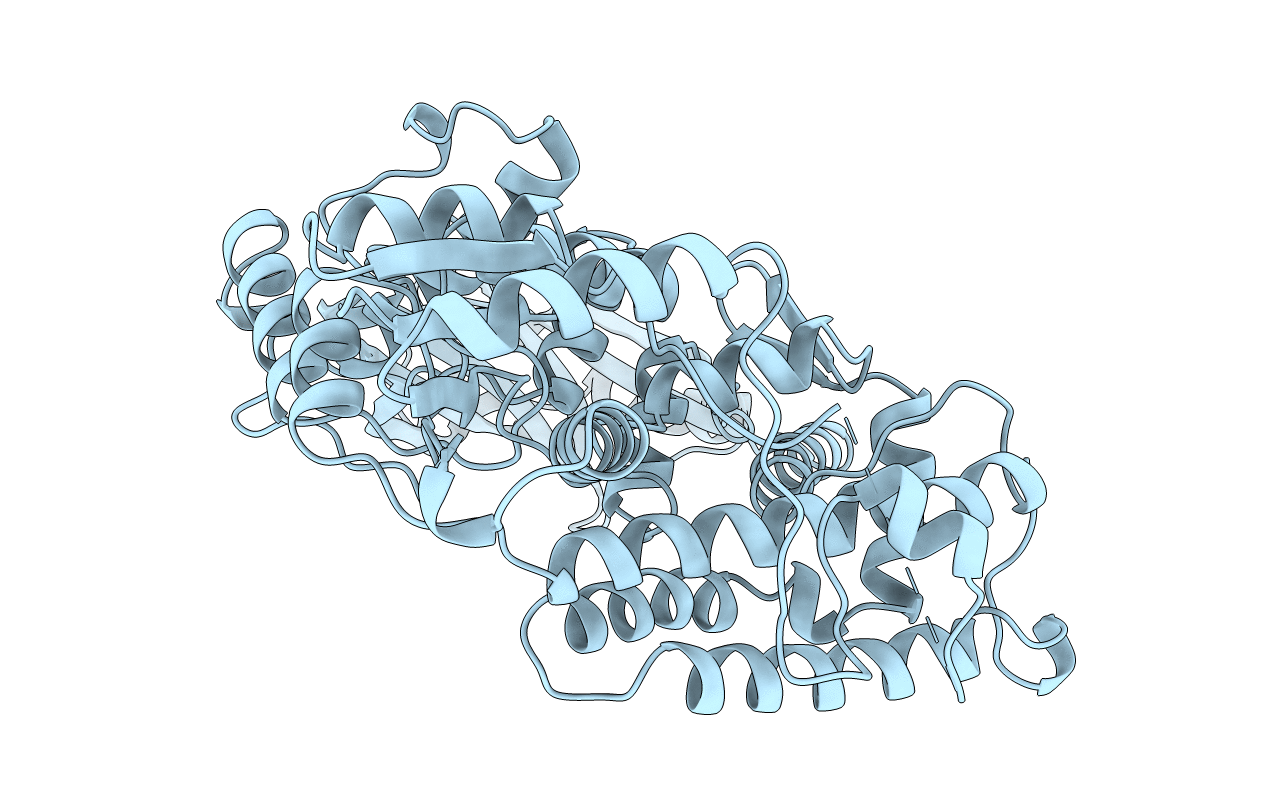
Title:
AMINO TERMINAL 67KDA DOMAIN OF ESCHERICHIA COLI DNA TOPOISOMERASE I (RESIDUES 2-590 OF MATURE PROTEIN) CLONING ARTIFACT ADDS TWO RESIDUES TO THE AMINO-TERMINUS WHICH WERE NOT OBSERVED IN THE EXPERIMENTAL ELECTRON DENSITY (GLY-2, SER-1).
Biological Source:
Source Organism(s):
Escherichia coli (Taxon ID: 562)
Method Details:
Experimental Method:
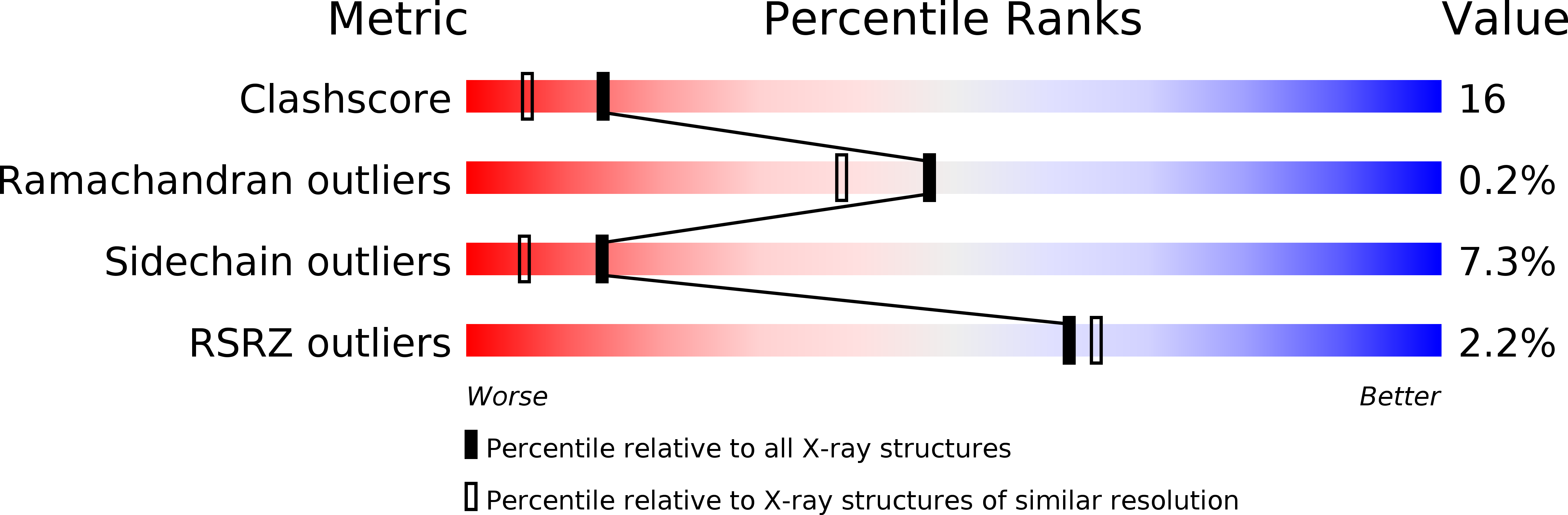
Resolution:
1.90 Å
R-Value Work:
0.21
R-Value Observed:
0.21
Space Group:
P 21 21 21