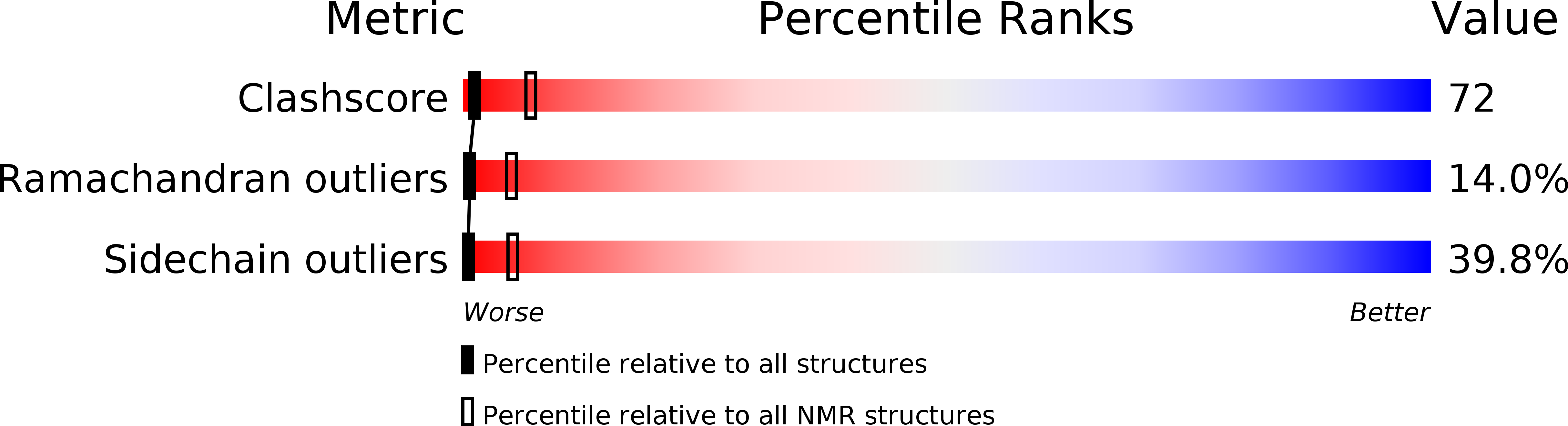
Deposition Date
2000-10-26
Release Date
2000-12-08
Last Version Date
2024-11-20
Entry Detail
PDB ID:
1E9T
Keywords:
Title:
High resolution solution structure of human intestinal trefoil factor
Biological Source:
Source Organism:
HOMO SAPIENS (Taxon ID: 9606)
Host Organism:
Method Details:
Experimental Method:
Conformers Calculated:
100
Conformers Submitted:
85
Selection Criteria:
CONSISTENCY WITH THE NMR STRUCTURAL DATA