Deposition Date
2000-06-14
Release Date
2001-06-14
Last Version Date
2025-12-17
Entry Detail
PDB ID:
1E3D
Keywords:
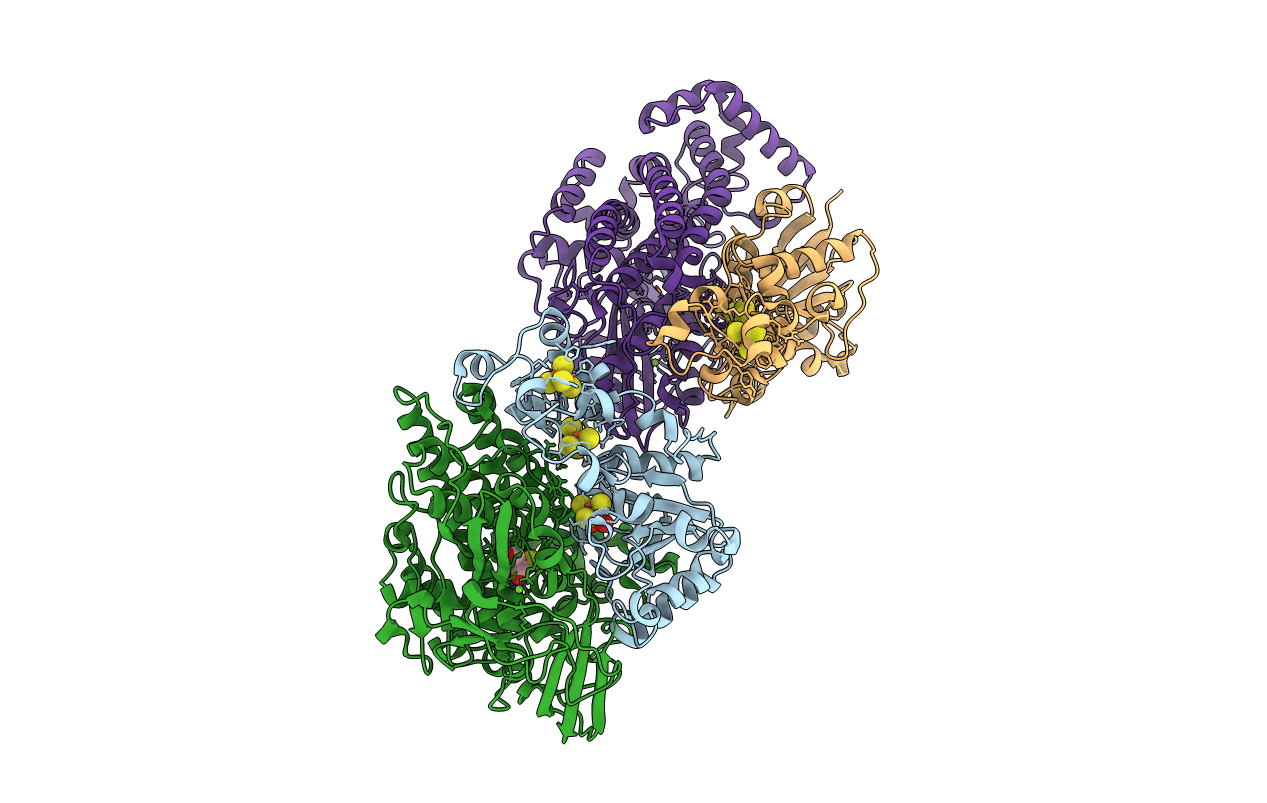
Title:
[NiFe] Hydrogenase from Desulfovibrio desulfuricans ATCC 27774
Biological Source:
Source Organism(s):
Desulfovibrio desulfuricans (Taxon ID: 876)
Method Details:
Experimental Method:
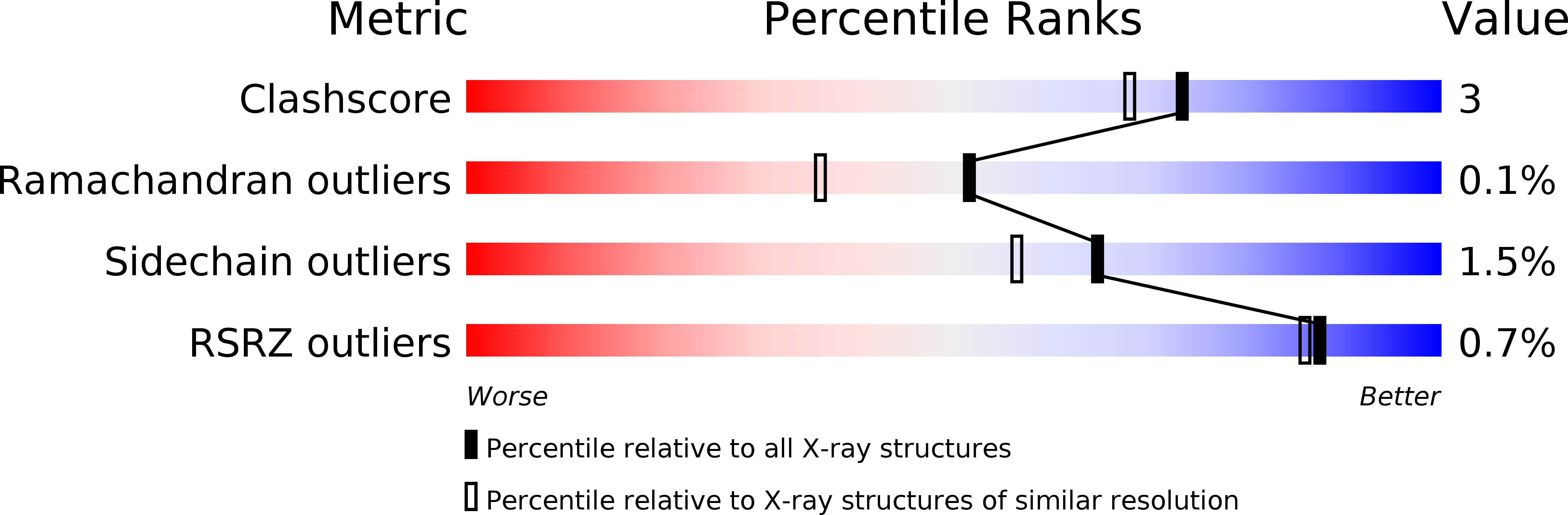
Resolution:
1.80 Å
R-Value Free:
0.22
R-Value Observed:
0.16
Space Group:
P 1 21 1