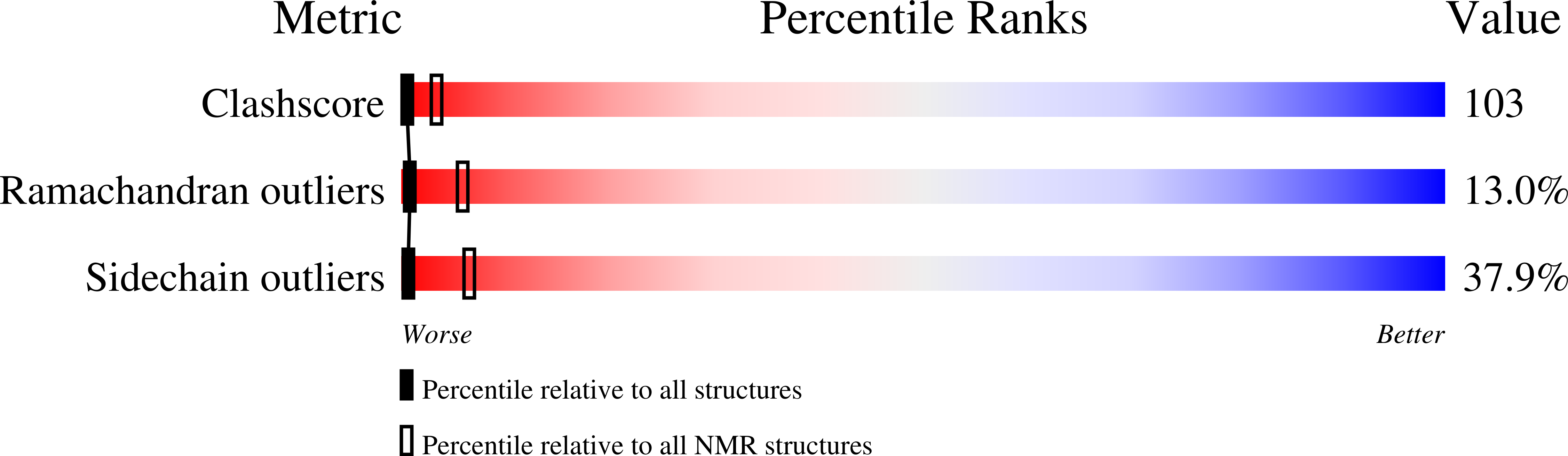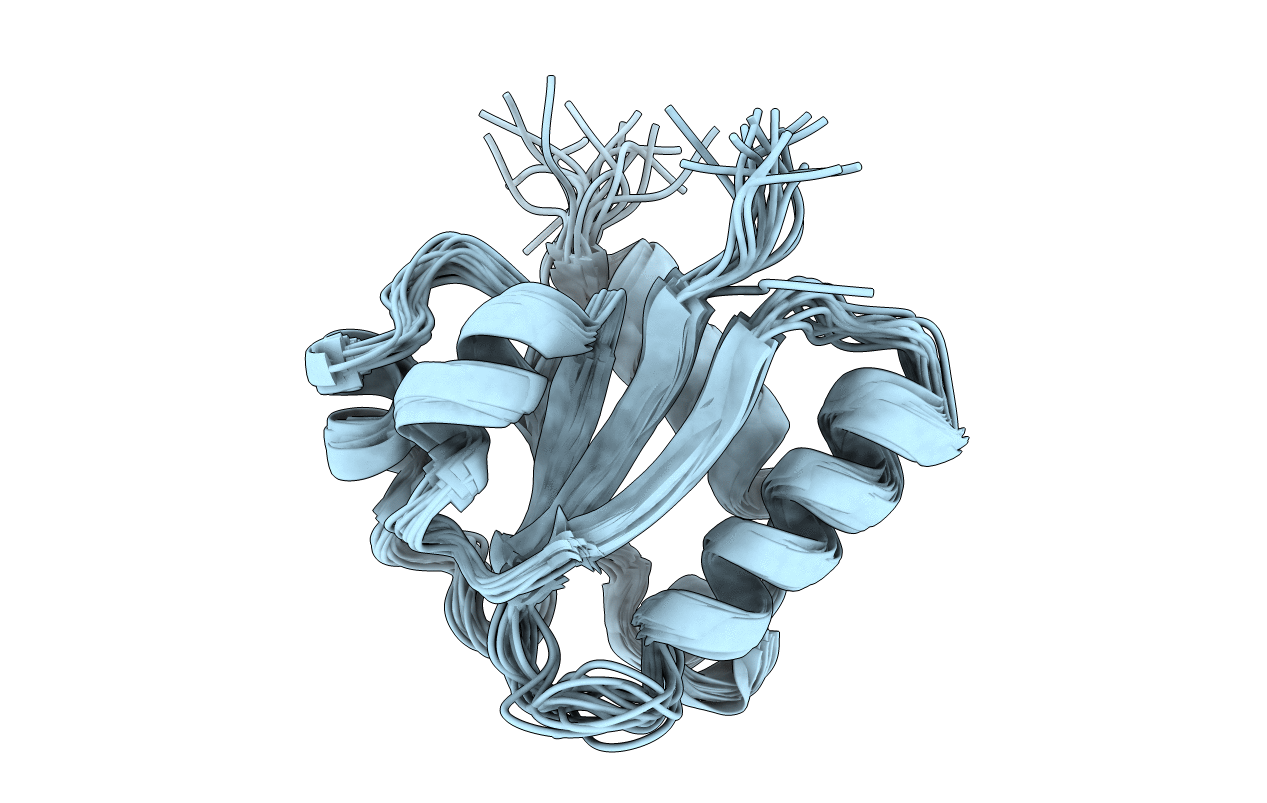
Deposition Date
1996-11-15
Release Date
1997-07-23
Last Version Date
2024-05-22
Entry Detail
PDB ID:
1E2B
Keywords:
Title:
NMR STRUCTURE OF THE C10S MUTANT OF ENZYME IIB CELLOBIOSE OF THE PHOSPHOENOL-PYRUVATE DEPENDENT PHOSPHOTRANSFERASE SYSTEM OF ESCHERICHIA COLI, 17 STRUCTURES
Biological Source:
Source Organism(s):
Escherichia coli (Taxon ID: 83333)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
32
Conformers Submitted:
17
Selection Criteria:
TARGET FUNCTION, NUMBER OF VIOLATIONS